Update upstream source from tag 'upstream/0.10.0'
Update to upstream version '0.10.0'
with Debian dir 56b5e009f1c2a96f88d664dc4d45b82749c898a9
Bas Couwenberg
2 years ago
| 6 | 6 | branches: [master] |
| 7 | 7 | schedule: |
| 8 | 8 | - cron: "0 0 * * *" |
| 9 | ||
| 10 | concurrency: | |
| 11 | group: ${{ github.workflow }}-${{ github.ref }} | |
| 12 | cancel-in-progress: true | |
| 9 | 13 | |
| 10 | 14 | jobs: |
| 11 | 15 | Linting: |
| 20 | 24 | needs: Linting |
| 21 | 25 | name: ${{ matrix.os }}, ${{ matrix.env }} |
| 22 | 26 | runs-on: ${{ matrix.os }} |
| 27 | defaults: | |
| 28 | run: | |
| 29 | shell: bash -l {0} | |
| 23 | 30 | strategy: |
| 31 | fail-fast: false | |
| 24 | 32 | matrix: |
| 25 | 33 | os: [ubuntu-latest] |
| 26 | 34 | postgis: [false] |
| 27 | 35 | dev: [false] |
| 28 | 36 | env: |
| 29 | - ci/envs/36-minimal.yaml | |
| 37 | - ci/envs/37-minimal.yaml | |
| 30 | 38 | - ci/envs/38-no-optional-deps.yaml |
| 31 | - ci/envs/36-pd025.yaml | |
| 39 | - ci/envs/37-pd10.yaml | |
| 32 | 40 | - ci/envs/37-latest-defaults.yaml |
| 33 | 41 | - ci/envs/37-latest-conda-forge.yaml |
| 34 | 42 | - ci/envs/38-latest-conda-forge.yaml |
| 58 | 66 | - uses: actions/checkout@v2 |
| 59 | 67 | |
| 60 | 68 | - name: Setup Conda |
| 61 | uses: s-weigand/setup-conda@v1 | |
| 69 | uses: conda-incubator/setup-miniconda@v2 | |
| 62 | 70 | with: |
| 63 | activate-conda: false | |
| 64 | ||
| 65 | - name: Install Env | |
| 66 | shell: bash | |
| 67 | run: conda env create -f ${{ matrix.env }} | |
| 71 | environment-file: ${{ matrix.env }} | |
| 68 | 72 | |
| 69 | 73 | - name: Check and Log Environment |
| 70 | shell: bash | |
| 71 | 74 | run: | |
| 72 | source activate test | |
| 73 | 75 | python -V |
| 74 | 76 | python -c "import geopandas; geopandas.show_versions();" |
| 75 | 77 | conda info |
| 86 | 88 | fi |
| 87 | 89 | |
| 88 | 90 | - name: Test without PyGEOS |
| 89 | shell: bash | |
| 90 | 91 | env: |
| 91 | 92 | USE_PYGEOS: 0 |
| 92 | 93 | run: | |
| 93 | source activate test | |
| 94 | 94 | pytest -v -r s -n auto --color=yes --cov=geopandas --cov-append --cov-report term-missing --cov-report xml geopandas/ |
| 95 | 95 | |
| 96 | 96 | - name: Test with PyGEOS |
| 97 | shell: bash | |
| 98 | 97 | if: env.HAS_PYGEOS == 1 |
| 99 | 98 | env: |
| 100 | 99 | USE_PYGEOS: 1 |
| 101 | 100 | run: | |
| 102 | source activate test | |
| 103 | 101 | pytest -v -r s -n auto --color=yes --cov=geopandas --cov-append --cov-report term-missing --cov-report xml geopandas/ |
| 104 | 102 | |
| 105 | 103 | - name: Test with PostGIS |
| 106 | shell: bash | |
| 107 | 104 | if: contains(matrix.env, '38-latest-conda-forge.yaml') && contains(matrix.os, 'ubuntu') |
| 108 | 105 | env: |
| 109 | 106 | PGUSER: postgres |
| 110 | 107 | PGPASSWORD: postgres |
| 111 | 108 | PGHOST: "127.0.0.1" |
| 109 | PGPORT: 5432 | |
| 112 | 110 | run: | |
| 113 | source activate test | |
| 114 | 111 | conda install postgis -c conda-forge |
| 115 | source ci/envs/setup_postgres.sh | |
| 112 | sh ci/scripts/setup_postgres.sh | |
| 116 | 113 | pytest -v -r s --color=yes --cov=geopandas --cov-append --cov-report term-missing --cov-report xml geopandas/io/tests/test_sql.py | tee /dev/stderr | if grep SKIPPED >/dev/null;then echo "TESTS SKIPPED, FAILING" && exit 1;fi |
| 117 | 114 | |
| 118 | 115 | - name: Test docstrings |
| 119 | shell: bash | |
| 120 | 116 | if: contains(matrix.env, '38-latest-conda-forge.yaml') && contains(matrix.os, 'ubuntu') |
| 121 | 117 | env: |
| 122 | 118 | USE_PYGEOS: 1 |
| 123 | 119 | run: | |
| 124 | source activate test | |
| 125 | 120 | pytest -v --color=yes --doctest-only geopandas --ignore=geopandas/datasets |
| 126 | 121 | |
| 127 | 122 | - uses: codecov/codecov-action@v1 |
| 0 | 0 | Changelog |
| 1 | 1 | ========= |
| 2 | ||
| 3 | Version 0.10.0 (October 3, 2021) | |
| 4 | -------------------------------- | |
| 5 | ||
| 6 | Highlights of this release: | |
| 7 | ||
| 8 | - A new `sjoin_nearest()` method to join based on proximity, with the | |
| 9 | ability to set a maximum search radius (#1865). In addition, the `sindex` | |
| 10 | attribute gained a new method for a "nearest" spatial index query (#1865, | |
| 11 | #2053). | |
| 12 | - A new `explore()` method on GeoDataFrame and GeoSeries with native support | |
| 13 | for interactive visualization based on folium / leaflet.js (#1953) | |
| 14 | - The `geopandas.sjoin()`/`overlay()`/`clip()` functions are now also | |
| 15 | available as methods on the GeoDataFrame (#2141, #1984, #2150). | |
| 16 | ||
| 17 | New features and improvements: | |
| 18 | ||
| 19 | - Add support for pandas' `value_counts()` method for geometry dtype (#2047). | |
| 20 | - The `explode()` method has a new `ignore_index` keyword (consistent with | |
| 21 | pandas' explode method) to reset the index in the result, and a new | |
| 22 | `index_parts` keywords to control whether a cumulative count indexing the | |
| 23 | parts of the exploded multi-geometries should be added (#1871). | |
| 24 | - `points_from_xy()` is now available as a GeoSeries method `from_xy` (#1936). | |
| 25 | - The `to_file()` method will now attempt to detect the driver (if not | |
| 26 | specified) based on the extension of the provided filename, instead of | |
| 27 | defaulting to ESRI Shapefile (#1609). | |
| 28 | - Support for the `storage_options` keyword in `read_parquet()` for | |
| 29 | specifying filesystem-specific options (e.g. for S3) based on fsspec (#2107). | |
| 30 | - The read/write functions now support `~` (user home directory) expansion (#1876). | |
| 31 | - Support the `convert_dtypes()` method from pandas to preserve the | |
| 32 | GeoDataFrame class (#2115). | |
| 33 | - Support WKB values in the hex format in `GeoSeries.from_wkb()` (#2106). | |
| 34 | - Update the `estimate_utm_crs()` method to handle crossing the antimeridian | |
| 35 | with pyproj 3.1+ (#2049). | |
| 36 | - Improved heuristic to decide how many decimals to show in the repr based on | |
| 37 | whether the CRS is projected or geographic (#1895). | |
| 38 | - Switched the default for `geocode()` from GeoCode.Farm to the Photon | |
| 39 | geocoding API (https://photon.komoot.io) (#2007). | |
| 40 | ||
| 41 | Deprecations and compatibility notes: | |
| 42 | ||
| 43 | - The `op=` keyword of `sjoin()` to indicate which spatial predicate to use | |
| 44 | for joining is being deprecated and renamed in favor of a new `predicate=` | |
| 45 | keyword (#1626). | |
| 46 | - The `cascaded_union` attribute is deprecated, use `unary_union` instead (#2074). | |
| 47 | - Constructing a GeoDataFrame with a duplicated "geometry" column is now | |
| 48 | disallowed. This can also raise an error in the `pd.concat(.., axis=1)` | |
| 49 | function if this results in duplicated active geometry columns (#2046). | |
| 50 | - The `explode()` method currently returns a GeoSeries/GeoDataFrame with a | |
| 51 | MultiIndex, with an additional level with indices of the parts of the | |
| 52 | exploded multi-geometries. For consistency with pandas, this will change in | |
| 53 | the future and the new `index_parts` keyword is added to control this. | |
| 54 | ||
| 55 | Bug fixes: | |
| 56 | ||
| 57 | - Fix in the `clip()` function to correctly clip MultiPoints instead of | |
| 58 | leaving them intact when partly outside of the clip bounds (#2148). | |
| 59 | - Fix `GeoSeries.isna()` to correctly return a boolean Series in case of an | |
| 60 | empty GeoSeries (#2073). | |
| 61 | - Fix the GeoDataFrame constructor to preserve the geometry name when the | |
| 62 | argument is already a GeoDataFrame object (i.e. `GeoDataFrame(gdf)`) (#2138). | |
| 63 | - Fix loss of the values' CRS when setting those values as a column | |
| 64 | (`GeoDataFrame.__setitem__`) (#1963) | |
| 65 | - Fix in `GeoDataFrame.apply()` to preserve the active geometry column name | |
| 66 | (#1955). | |
| 67 | - Fix in `sjoin()` to not ignore the suffixes in case of a right-join | |
| 68 | (`how="right`) (#2065). | |
| 69 | - Fix `GeoDataFrame.explode()` with a MultiIndex (#1945). | |
| 70 | - Fix the handling of missing values in `to/from_wkb` and `to_from_wkt` (#1891). | |
| 71 | - Fix `to_file()` and `to_json()` when DataFrame has duplicate columns to | |
| 72 | raise an error (#1900). | |
| 73 | - Fix bug in the colors shown with user-defined classification scheme (#2019). | |
| 74 | - Fix handling of the `path_effects` keyword in `plot()` (#2127). | |
| 75 | - Fix `GeoDataFrame.explode()` to preserve `attrs` (#1935) | |
| 76 | ||
| 77 | Notes on (optional) dependencies: | |
| 78 | ||
| 79 | - GeoPandas 0.9.0 dropped support for Python 3.6 and pandas 0.24. Further, | |
| 80 | the minimum required versions are numpy 1.18, shapely 1.6, fiona 1.8, | |
| 81 | matplotlib 3.1 and pyproj 2.2. | |
| 82 | - Plotting with a classification schema now requires mapclassify version >= | |
| 83 | 2.4 (#1737). | |
| 84 | - Compatibility fixes for the latest numpy in combination with Shapely 1.7 (#2072) | |
| 85 | - Compatibility fixes for the upcoming Shapely 1.8 (#2087). | |
| 86 | - Compatibility fixes for the latest PyGEOS (#1872, #2014) and matplotlib | |
| 87 | (colorbar issue, #2066). | |
| 88 | ||
| 2 | 89 | |
| 3 | 90 | Version 0.9.0 (February 28, 2021) |
| 4 | 91 | --------------------------------- |
| 77 | 164 | - Fix regression in the `plot()` method raising an error with empty |
| 78 | 165 | geometries (#1702, #1828). |
| 79 | 166 | - Fix `geopandas.overlay()` to preserve geometries of the correct type which |
| 80 | are nested withing a GeometryCollection as a result of the overlay | |
| 167 | are nested within a GeometryCollection as a result of the overlay | |
| 81 | 168 | operation (#1582). In addition, a warning will now be raised if geometries |
| 82 | 169 | of different type are dropped from the result (#1554). |
| 83 | 170 | - Fix the repr of an empty GeoSeries to not show spurious warnings (#1673). |
| 84 | 171 | - Fix the `.crs` for empty GeoDataFrames (#1560). |
| 85 | - Fix `geopandas.clip` to preserve the correct geometry column name (#1566). | |
| 172 | - Fix `geopandas.clip` to preserve the correct geometry column name (#1566). | |
| 86 | 173 | - Fix bug in `plot()` method when using `legend_kwds` with multiple subplots |
| 87 | 174 | (#1583) |
| 88 | 175 | - Fix spurious warning with `missing_kwds` keyword of the `plot()` method |
| 148 | 235 | New features and improvements: |
| 149 | 236 | |
| 150 | 237 | - IO enhancements: |
| 238 | ||
| 151 | 239 | - New `GeoDataFrame.to_postgis()` method to write to PostGIS database (#1248). |
| 152 | 240 | - New Apache Parquet and Feather file format support (#1180, #1435) |
| 153 | 241 | - Allow appending to files with `GeoDataFrame.to_file` (#1229). |
| 156 | 244 | returned (#1383). |
| 157 | 245 | - `geopandas.read_file` now supports reading from file-like objects (#1329). |
| 158 | 246 | - `GeoDataFrame.to_file` now supports specifying the CRS to write to the file |
| 159 | (#802). By default it still uses the CRS of the GeoDataFrame. | |
| 247 | (#802). By default it still uses the CRS of the GeoDataFrame. | |
| 160 | 248 | - New `chunksize` keyword in `geopandas.read_postgis` to read a query in |
| 161 | 249 | chunks (#1123). |
| 250 | ||
| 162 | 251 | - Improvements related to geometry columns and CRS: |
| 252 | ||
| 163 | 253 | - Any column of the GeoDataFrame that has a "geometry" dtype is now returned |
| 164 | 254 | as a GeoSeries. This means that when having multiple geometry columns, not |
| 165 | 255 | only the "active" geometry column is returned as a GeoSeries, but also |
| 171 | 261 | from the column itself (eg `gdf["other_geom_column"].crs`) (#1339). |
| 172 | 262 | - New `set_crs()` method on GeoDataFrame/GeoSeries to set the CRS of naive |
| 173 | 263 | geometries (#747). |
| 264 | ||
| 174 | 265 | - Improvements related to plotting: |
| 266 | ||
| 175 | 267 | - The y-axis is now scaled depending on the center of the plot when using a |
| 176 | 268 | geographic CRS, instead of using an equal aspect ratio (#1290). |
| 177 | 269 | - When passing a column of categorical dtype to the `column=` keyword of the |
| 182 | 274 | `legend_kwds` accept two new keywords to control the formatting of the |
| 183 | 275 | legend: `fmt` with a format string for the bin edges (#1253), and `labels` |
| 184 | 276 | to pass fully custom class labels (#1302). |
| 277 | ||
| 185 | 278 | - New `covers()` and `covered_by()` methods on GeoSeries/GeoDataframe for the |
| 186 | 279 | equivalent spatial predicates (#1460, #1462). |
| 187 | 280 | - GeoPandas now warns when using distance-based methods with data in a |
| 193 | 286 | CRS, a deprecation warning is raised when both CRS don't match, and in the |
| 194 | 287 | future an error will be raised in such a case. You can use the new `set_crs` |
| 195 | 288 | method to override an existing CRS. See |
| 196 | [the docs](https://geopandas.readthedocs.io/en/latest/projections.html#projection-for-multiple-geometry-columns). | |
| 289 | [the docs](https://geopandas.readthedocs.io/en/latest/projections.html#projection-for-multiple-geometry-columns). | |
| 197 | 290 | - The helper functions in the `geopandas.plotting` module are deprecated for |
| 198 | 291 | public usage (#656). |
| 199 | 292 | - The `geopandas.io` functions are deprecated, use the top-level `read_file` and |
| 314 | 407 | API changes: |
| 315 | 408 | |
| 316 | 409 | - A refactor of the internals based on the pandas ExtensionArray interface (#1000). The main user visible changes are: |
| 410 | ||
| 317 | 411 | - The `.dtype` of a GeoSeries is now a `'geometry'` dtype (and no longer a numpy `object` dtype). |
| 318 | 412 | - The `.values` of a GeoSeries now returns a custom `GeometryArray`, and no longer a numpy array. To get back a numpy array of Shapely scalars, you can convert explicitly using `np.asarray(..)`. |
| 413 | ||
| 319 | 414 | - The `GeoSeries` constructor now raises a warning when passed non-geometry data. Currently the constructor falls back to return a pandas `Series`, but in the future this will raise an error (#1085). |
| 320 | 415 | - The missing value handling has been changed to now separate the concepts of missing geometries and empty geometries (#601, 1062). In practice this means that (see [the docs](https://geopandas.readthedocs.io/en/v0.6.0/missing_empty.html) for more details): |
| 416 | ||
| 321 | 417 | - `GeoSeries.isna` now considers only missing values, and if you want to check for empty geometries, you can use `GeoSeries.is_empty` (`GeoDataFrame.isna` already only looked at missing values). |
| 322 | 418 | - `GeoSeries.dropna` now actually drops missing values (before it didn't drop either missing or empty geometries) |
| 323 | 419 | - `GeoSeries.fillna` only fills missing values (behaviour unchanged). |
| 360 | 456 | * Significant performance improvement (around 10x) for `GeoDataFrame.iterfeatures`, |
| 361 | 457 | which also improves `GeoDataFrame.to_file` (#864). |
| 362 | 458 | * File IO enhancements based on Fiona 1.8: |
| 459 | ||
| 363 | 460 | * Support for writing bool dtype (#855) and datetime dtype, if the file format supports it (#728). |
| 364 | 461 | * Support for writing dataframes with multiple geometry types, if the file format allows it (e.g. GeoJSON for all types, or ESRI Shapefile for Polygon+MultiPolygon) (#827, #867, #870). |
| 462 | ||
| 365 | 463 | * Compatibility with pyproj >= 2 (#962). |
| 366 | 464 | * A new `geopandas.points_from_xy()` helper function to convert x and y coordinates to Point objects (#896). |
| 367 | * The `buffer` and `interpolate` methods now accept an array-like to specify a variable distance for each geometry (#781). | |
| 465 | * The `buffer` and `interpolate` methods now accept an array-like to specify a variable distance for each geometry (#781). | |
| 368 | 466 | * Addition of a `relate` method, corresponding to the shapely method that returns the DE-9IM matrix (#853). |
| 369 | 467 | * Plotting improvements: |
| 468 | ||
| 370 | 469 | * Performance improvement in plotting by only flattening the geometries if there are actually 'Multi' geometries (#785). |
| 371 | 470 | * Choropleths: access to all `mapclassify` classification schemes and addition of the `classification_kwds` keyword in the `plot` method to specify options for the scheme (#876). |
| 372 | 471 | * Ability to specify a matplotlib axes object on which to plot the color bar with the `cax` keyword, in order to have more control over the color bar placement (#894). |
| 472 | ||
| 373 | 473 | * Changed the default provider in ``geopandas.tools.geocode`` from Google (now requires an API key) to Geocode.Farm (#907, #975). |
| 374 | 474 | |
| 375 | 475 | Bug fixes: |
| 413 | 513 | * Permit setting markersize for Point GeoSeries plots with column values (#633) |
| 414 | 514 | * Started an example gallery (#463, #690, #717) |
| 415 | 515 | * Support for plotting MultiPoints (#683) |
| 416 | * Testing functionalty (e.g. `assert_geodataframe_equal`) is now publicly exposed (#707) | |
| 516 | * Testing functionality (e.g. `assert_geodataframe_equal`) is now publicly exposed (#707) | |
| 417 | 517 | * Add `explode` method to GeoDataFrame (similar to the GeoSeries method) (#671) |
| 418 | 518 | * Set equal aspect on active axis on multi-axis figures (#718) |
| 419 | 519 | * Pass array of values to column argument in `plot` (#770) |
| 18 | 18 | - Install the requirements for the development environment (one can do this |
| 19 | 19 | with either conda, and the environment.yml file, or pip, and the |
| 20 | 20 | requirements-dev.txt file, and can use the pandas contributing guidelines |
| 21 | as a guide). | |
| 21 | as a guide). | |
| 22 | 22 | - All existing tests should pass. Please make sure that the test |
| 23 | 23 | suite passes, both locally and on |
| 24 | 24 | [GitHub Actions](https://github.com/geopandas/geopandas/actions). Status on |
| 38 | 38 | Style |
| 39 | 39 | ----- |
| 40 | 40 | |
| 41 | - GeoPandas supports Python 3.6+ only. The last version of GeoPandas | |
| 41 | - GeoPandas supports Python 3.7+ only. The last version of GeoPandas | |
| 42 | 42 | supporting Python 2 is 0.6. |
| 43 | 43 | |
| 44 | 44 | - GeoPandas follows [the PEP 8 |
| 0 | 0 | from geopandas import GeoDataFrame, GeoSeries, read_file, datasets, overlay |
| 1 | from shapely.geometry import Polygon | |
| 1 | import numpy as np | |
| 2 | from shapely.geometry import Point, Polygon | |
| 2 | 3 | |
| 3 | 4 | |
| 4 | 5 | class Countries: |
| 5 | 6 | |
| 6 | param_names = ['op'] | |
| 7 | param_names = ['how'] | |
| 7 | 8 | params = [('intersection', 'union', 'identity', 'symmetric_difference', |
| 8 | 9 | 'difference')] |
| 9 | 10 | |
| 19 | 20 | self.countries = countries |
| 20 | 21 | self.capitals = capitals |
| 21 | 22 | |
| 22 | def time_overlay(self, op): | |
| 23 | overlay(self.countries, self.capitals, how=op) | |
| 23 | def time_overlay(self, how): | |
| 24 | overlay(self.countries, self.capitals, how=how) | |
| 24 | 25 | |
| 25 | 26 | |
| 26 | 27 | class Small: |
| 27 | 28 | |
| 28 | param_names = ['op'] | |
| 29 | param_names = ['how'] | |
| 29 | 30 | params = [('intersection', 'union', 'identity', 'symmetric_difference', |
| 30 | 31 | 'difference')] |
| 31 | 32 | |
| 40 | 41 | |
| 41 | 42 | self.df1, self.df2 = df1, df2 |
| 42 | 43 | |
| 43 | def time_overlay(self, op): | |
| 44 | overlay(self.df1, self.df2, how=op) | |
| 44 | def time_overlay(self, how): | |
| 45 | overlay(self.df1, self.df2, how=how) | |
| 46 | ||
| 47 | ||
| 48 | class ManyPoints: | |
| 49 | ||
| 50 | param_names = ['how'] | |
| 51 | params = [('intersection', 'union', 'identity', 'symmetric_difference', | |
| 52 | 'difference')] | |
| 53 | ||
| 54 | def setup(self, *args): | |
| 55 | ||
| 56 | points = GeoDataFrame(geometry=[Point(i, i) for i in range(1000)]) | |
| 57 | base = np.array([[0, 0], [0, 100], [100, 100], [100, 0]]) | |
| 58 | polys = GeoDataFrame( | |
| 59 | geometry=[Polygon(base + i * 100) for i in range(10)]) | |
| 60 | ||
| 61 | self.df1, self.df2 = points, polys | |
| 62 | ||
| 63 | def time_overlay(self, how): | |
| 64 | overlay(self.df1, self.df2, how=how) | |
| 6 | 6 | |
| 7 | 7 | class Bench: |
| 8 | 8 | |
| 9 | param_names = ['geom_type'] | |
| 10 | params = [('Point', 'LineString', 'Polygon', 'MultiPolygon', 'mixed')] | |
| 9 | param_names = ["geom_type"] | |
| 10 | params = [("Point", "LineString", "Polygon", "MultiPolygon", "mixed")] | |
| 11 | 11 | |
| 12 | 12 | def setup(self, geom_type): |
| 13 | 13 | |
| 14 | if geom_type == 'Point': | |
| 14 | if geom_type == "Point": | |
| 15 | 15 | geoms = GeoSeries([Point(i, i) for i in range(1000)]) |
| 16 | elif geom_type == 'LineString': | |
| 17 | geoms = GeoSeries([LineString([(random.random(), random.random()) | |
| 18 | for _ in range(5)]) | |
| 19 | for _ in range(100)]) | |
| 20 | elif geom_type == 'Polygon': | |
| 21 | geoms = GeoSeries([Polygon([(random.random(), random.random()) | |
| 22 | for _ in range(3)]) | |
| 23 | for _ in range(100)]) | |
| 24 | elif geom_type == 'MultiPolygon': | |
| 16 | elif geom_type == "LineString": | |
| 25 | 17 | geoms = GeoSeries( |
| 26 | [MultiPolygon([Polygon([(random.random(), random.random()) | |
| 27 | for _ in range(3)]) | |
| 28 | for _ in range(3)]) | |
| 29 | for _ in range(20)]) | |
| 30 | elif geom_type == 'mixed': | |
| 18 | [ | |
| 19 | LineString([(random.random(), random.random()) for _ in range(5)]) | |
| 20 | for _ in range(100) | |
| 21 | ] | |
| 22 | ) | |
| 23 | elif geom_type == "Polygon": | |
| 24 | geoms = GeoSeries( | |
| 25 | [ | |
| 26 | Polygon([(random.random(), random.random()) for _ in range(3)]) | |
| 27 | for _ in range(100) | |
| 28 | ] | |
| 29 | ) | |
| 30 | elif geom_type == "MultiPolygon": | |
| 31 | geoms = GeoSeries( | |
| 32 | [ | |
| 33 | MultiPolygon( | |
| 34 | [ | |
| 35 | Polygon( | |
| 36 | [(random.random(), random.random()) for _ in range(3)] | |
| 37 | ) | |
| 38 | for _ in range(3) | |
| 39 | ] | |
| 40 | ) | |
| 41 | for _ in range(20) | |
| 42 | ] | |
| 43 | ) | |
| 44 | elif geom_type == "mixed": | |
| 31 | 45 | g1 = GeoSeries([Point(i, i) for i in range(100)]) |
| 32 | g2 = GeoSeries([LineString([(random.random(), random.random()) | |
| 33 | for _ in range(5)]) | |
| 34 | for _ in range(100)]) | |
| 35 | g3 = GeoSeries([Polygon([(random.random(), random.random()) | |
| 36 | for _ in range(3)]) | |
| 37 | for _ in range(100)]) | |
| 46 | g2 = GeoSeries( | |
| 47 | [ | |
| 48 | LineString([(random.random(), random.random()) for _ in range(5)]) | |
| 49 | for _ in range(100) | |
| 50 | ] | |
| 51 | ) | |
| 52 | g3 = GeoSeries( | |
| 53 | [ | |
| 54 | Polygon([(random.random(), random.random()) for _ in range(3)]) | |
| 55 | for _ in range(100) | |
| 56 | ] | |
| 57 | ) | |
| 38 | 58 | |
| 39 | 59 | geoms = g1 |
| 40 | geoms.iloc[np.random.randint(0, 100, 50)] = g2 | |
| 41 | geoms.iloc[np.random.randint(0, 100, 33)] = g3 | |
| 60 | geoms.iloc[np.random.randint(0, 100, 50)] = g2.iloc[:50] | |
| 61 | geoms.iloc[np.random.randint(0, 100, 33)] = g3.iloc[:33] | |
| 42 | 62 | |
| 43 | 63 | print(geoms.geom_type.value_counts()) |
| 44 | 64 | |
| 45 | df = GeoDataFrame({'geometry': geoms, | |
| 46 | 'values': np.random.randn(len(geoms))}) | |
| 65 | df = GeoDataFrame({"geometry": geoms, "values": np.random.randn(len(geoms))}) | |
| 47 | 66 | |
| 48 | 67 | self.geoms = geoms |
| 49 | 68 | self.df = df |
| 52 | 71 | self.geoms.plot() |
| 53 | 72 | |
| 54 | 73 | def time_plot_values(self, *args): |
| 55 | self.df.plot(column='values') | |
| 56 | ||
| 74 | self.df.plot(column="values") | |
| 0 | name: test | |
| 1 | channels: | |
| 2 | - defaults | |
| 3 | - conda-forge | |
| 4 | dependencies: | |
| 5 | - python=3.6 | |
| 6 | # required | |
| 7 | - numpy=1.15 | |
| 8 | - pandas==0.24 | |
| 9 | - shapely=1.6 | |
| 10 | - fiona=1.8.13 | |
| 11 | #- pyproj | |
| 12 | # testing | |
| 13 | - pytest | |
| 14 | - pytest-cov | |
| 15 | - pytest-xdist | |
| 16 | - fsspec | |
| 17 | # optional | |
| 18 | - rtree | |
| 19 | - matplotlib | |
| 20 | - matplotlib=2.2 | |
| 21 | - mapclassify>=2.2.0 | |
| 22 | - geopy | |
| 23 | - SQLalchemy | |
| 24 | - libspatialite | |
| 25 | - pyarrow | |
| 26 | - pip: | |
| 27 | - pyproj==2.2.2 |
| 0 | name: test | |
| 1 | channels: | |
| 2 | - defaults | |
| 3 | dependencies: | |
| 4 | - python=3.6 | |
| 5 | # required | |
| 6 | - pandas=0.25 | |
| 7 | - shapely | |
| 8 | - fiona | |
| 9 | #- pyproj | |
| 10 | - geos | |
| 11 | # testing | |
| 12 | - pytest | |
| 13 | - pytest-cov | |
| 14 | - pytest-xdist | |
| 15 | - fsspec | |
| 16 | # optional | |
| 17 | - rtree | |
| 18 | - matplotlib | |
| 19 | #- geopy | |
| 20 | - SQLalchemy | |
| 21 | - libspatialite | |
| 22 | - pyarrow | |
| 23 | - pip: | |
| 24 | - pyproj==2.3.1 | |
| 25 | - geopy | |
| 26 | - mapclassify==2.2.0 |
| 17 | 17 | - rtree |
| 18 | 18 | - matplotlib |
| 19 | 19 | - mapclassify |
| 20 | - folium | |
| 21 | - xyzservices | |
| 20 | 22 | - scipy |
| 21 | 23 | - geopy |
| 22 | 24 | - SQLalchemy |
| 23 | 25 | - libspatialite |
| 24 | 26 | - pyarrow |
| 25 | ||
| 27 |
| 1 | 1 | channels: |
| 2 | 2 | - defaults |
| 3 | 3 | dependencies: |
| 4 | - python=3.7.3 | |
| 4 | - python=3.7 | |
| 5 | 5 | # required |
| 6 | 6 | - pandas |
| 7 | 7 | - shapely |
| 19 | 19 | #- geopy |
| 20 | 20 | - SQLalchemy |
| 21 | 21 | - libspatialite |
| 22 | - pyarrow | |
| 23 | 22 | - pip: |
| 24 | 23 | - geopy |
| 25 | 24 | - mapclassify |
| 25 | - pyarrow | |
| 26 | - folium | |
| 27 | - xyzservices | |
| 0 | name: test | |
| 1 | channels: | |
| 2 | - defaults | |
| 3 | - conda-forge | |
| 4 | dependencies: | |
| 5 | - python=3.7 | |
| 6 | # required | |
| 7 | - numpy=1.18 | |
| 8 | - pandas==0.25 | |
| 9 | - shapely=1.6 | |
| 10 | - fiona=1.8.13 | |
| 11 | #- pyproj | |
| 12 | # testing | |
| 13 | - pytest | |
| 14 | - pytest-cov | |
| 15 | - pytest-xdist | |
| 16 | - fsspec | |
| 17 | # optional | |
| 18 | - rtree | |
| 19 | - matplotlib | |
| 20 | - matplotlib=3.1 | |
| 21 | # - mapclassify=2.4.0 - doesn't build due to conflicts | |
| 22 | - geopy | |
| 23 | - SQLalchemy | |
| 24 | - libspatialite | |
| 25 | - pyarrow | |
| 26 | - pip: | |
| 27 | - pyproj==2.2.2 |
| 0 | name: test | |
| 1 | channels: | |
| 2 | - defaults | |
| 3 | dependencies: | |
| 4 | - python=3.7 | |
| 5 | # required | |
| 6 | - pandas=1.0 | |
| 7 | - shapely | |
| 8 | - fiona | |
| 9 | - numpy=<1.19 | |
| 10 | #- pyproj | |
| 11 | - geos | |
| 12 | # testing | |
| 13 | - pytest | |
| 14 | - pytest-cov | |
| 15 | - pytest-xdist | |
| 16 | - fsspec | |
| 17 | # optional | |
| 18 | - rtree | |
| 19 | - matplotlib | |
| 20 | #- geopy | |
| 21 | - SQLalchemy | |
| 22 | - libspatialite | |
| 23 | - pip | |
| 24 | - pip: | |
| 25 | - pyproj==3.0.1 | |
| 26 | - geopy | |
| 27 | - mapclassify==2.4.0 | |
| 28 | - pyarrow |
| 20 | 20 | - pyarrow |
| 21 | 21 | - pip: |
| 22 | 22 | - geopy |
| 23 | - mapclassify>=2.2.0 | |
| 23 | - mapclassify>=2.4.0 | |
| 24 | 24 | # dev versions of packages |
| 25 | - git+https://github.com/numpy/numpy.git@master | |
| 25 | - git+https://github.com/numpy/numpy.git@main | |
| 26 | 26 | - git+https://github.com/pydata/pandas.git@master |
| 27 | 27 | - git+https://github.com/matplotlib/matplotlib.git@master |
| 28 | 28 | - git+https://github.com/Toblerity/Shapely.git@master |
| 29 | 29 | - git+https://github.com/pygeos/pygeos.git@master |
| 30 | - git+https://github.com/python-visualization/folium.git@master | |
| 31 | - git+https://github.com/geopandas/xyzservices.git@main | |
| 32 |
| 3 | 3 | dependencies: |
| 4 | 4 | - python=3.8 |
| 5 | 5 | # required |
| 6 | - pandas | |
| 6 | - pandas=1.3.2 # temporary pin because 1.3.3 has regression for overlay (GH2101) | |
| 7 | 7 | - shapely |
| 8 | 8 | - fiona |
| 9 | 9 | - pyproj |
| 17 | 17 | - rtree |
| 18 | 18 | - matplotlib |
| 19 | 19 | - mapclassify |
| 20 | - folium | |
| 21 | - xyzservices | |
| 20 | 22 | - scipy |
| 21 | 23 | - geopy |
| 22 | 24 | # installed in tests.yaml, because not available on windows |
| 16 | 16 | # optional |
| 17 | 17 | - rtree |
| 18 | 18 | - matplotlib |
| 19 | - descartes | |
| 20 | 19 | - mapclassify |
| 20 | - folium | |
| 21 | - xyzservices | |
| 21 | 22 | - scipy |
| 22 | 23 | - geopy |
| 23 | 24 | # installed in tests.yaml, because not available on windows |
| 29 | 30 | - pyarrow |
| 30 | 31 | # doctest testing |
| 31 | 32 | - pytest-doctestplus |
| 33 | ||
| 0 | #!/bin/bash -e | |
| 1 | ||
| 2 | echo "Setting up Postgresql" | |
| 3 | ||
| 4 | mkdir -p ${HOME}/var | |
| 5 | rm -rf ${HOME}/var/db | |
| 6 | ||
| 7 | pg_ctl initdb -D ${HOME}/var/db | |
| 8 | pg_ctl start -D ${HOME}/var/db | |
| 9 | ||
| 10 | echo -n 'waiting for postgres' | |
| 11 | while [ ! -e /tmp/.s.PGSQL.5432 ]; do | |
| 12 | sleep 1 | |
| 13 | echo -n '.' | |
| 14 | done | |
| 15 | ||
| 16 | createuser -U ${USER} -s postgres | |
| 17 | createdb --owner=postgres test_geopandas | |
| 18 | psql -d test_geopandas -q -c "CREATE EXTENSION postgis" | |
| 19 | ||
| 20 | echo "Done setting up Postgresql" |
| 0 | #!/bin/sh | |
| 1 | set -e | |
| 2 | ||
| 3 | if [ -z "${PGUSER}" ] || [ -z "${PGPORT}" ]; then | |
| 4 | echo "Environment variables PGUSER and PGPORT must be set" | |
| 5 | exit 1 | |
| 6 | fi | |
| 7 | ||
| 8 | PGDATA=$(mktemp -d /tmp/postgres.XXXXXX) | |
| 9 | echo "Setting up PostgreSQL in ${PGDATA} on port ${PGPORT}" | |
| 10 | ||
| 11 | pg_ctl -D ${PGDATA} initdb | |
| 12 | pg_ctl -D ${PGDATA} start | |
| 13 | ||
| 14 | SOCKETPATH="/tmp/.s.PGSQL.${PGPORT}" | |
| 15 | echo -n 'waiting for postgres' | |
| 16 | while [ ! -e ${SOCKETPATH} ]; do | |
| 17 | sleep 1 | |
| 18 | echo -n '.' | |
| 19 | done | |
| 20 | echo | |
| 21 | ||
| 22 | echo "Done setting up PostgreSQL. When finished, stop and cleanup using:" | |
| 23 | echo | |
| 24 | echo " pg_ctl -D ${PGDATA} stop" | |
| 25 | echo " rm -rf ${PGDATA}" | |
| 26 | echo | |
| 27 | ||
| 28 | createuser -U ${USER} -s ${PGUSER} | |
| 29 | createdb --owner=${PGUSER} test_geopandas | |
| 30 | psql -d test_geopandas -q -c "CREATE EXTENSION postgis" | |
| 31 | ||
| 32 | echo "PostGIS server ready." |
| 1 | 1 | channels: |
| 2 | 2 | - conda-forge |
| 3 | 3 | dependencies: |
| 4 | - python=3.9.1 | |
| 5 | - pandas=1.2.2 | |
| 4 | - python=3.9.7 | |
| 5 | - pandas=1.3.2 | |
| 6 | 6 | - shapely=1.7.1 |
| 7 | - fiona=1.8.18 | |
| 8 | - pyproj=3.0.0.post1 | |
| 7 | - fiona=1.8.20 | |
| 8 | - pyproj=3.2.1 | |
| 9 | 9 | - rtree=0.9.7 |
| 10 | - geopy=2.1.0 | |
| 11 | - matplotlib=3.3.4 | |
| 12 | - mapclassify=2.4.2 | |
| 13 | - sphinx=3.5.1 | |
| 14 | - pydata-sphinx-theme=0.4.3 | |
| 10 | - geopy=2.2.0 | |
| 11 | - matplotlib=3.4.3 | |
| 12 | - mapclassify=2.4.3 | |
| 13 | - sphinx=4.2.0 | |
| 14 | - pydata-sphinx-theme=0.6.3 | |
| 15 | 15 | - numpydoc=1.1.0 |
| 16 | - ipython=7.20.0 | |
| 17 | - pillow=8.1.0 | |
| 16 | - ipython=7.27.0 | |
| 17 | - pillow=8.3.2 | |
| 18 | 18 | - mock=4.0.3 |
| 19 | - cartopy=0.18.0 | |
| 19 | - cartopy=0.20.0 | |
| 20 | 20 | - pyepsg=0.4.0 |
| 21 | 21 | - contextily=1.1.0 |
| 22 | - rasterio=1.2.0 | |
| 23 | - geoplot=0.4.1 | |
| 24 | - sphinx-gallery=0.8.2 | |
| 25 | - jinja2=2.11.3 | |
| 22 | - rasterio=1.2.8 | |
| 23 | - geoplot=0.4.4 | |
| 24 | - sphinx-gallery=0.9.0 | |
| 25 | - jinja2=3.0.1 | |
| 26 | 26 | - doc2dash=2.3.0 |
| 27 | - matplotlib-scalebar=0.7.2 | |
| 27 | 28 | # specify additional dependencies to reduce solving for conda |
| 28 | - gdal=3.1.4 | |
| 29 | - libgdal=3.1.4 | |
| 30 | - proj=7.2.0 | |
| 31 | - geos=3.9.0 | |
| 32 | - nbsphinx=0.8.1 | |
| 33 | - jupyter_client=6.1.11 | |
| 34 | - ipykernel=5.4.3 | |
| 35 | - myst-parser=0.13.5 | |
| 29 | - gdal=3.3.2 | |
| 30 | - libgdal=3.3.2 | |
| 31 | - proj=8.0.1 | |
| 32 | - geos=3.9.1 | |
| 33 | - nbsphinx=0.8.7 | |
| 34 | - jupyter_client=7.0.3 | |
| 35 | - ipykernel=6.4.1 | |
| 36 | - myst-parser=0.15.2 | |
| 36 | 37 | - folium=0.12.0 |
| 37 | - libpysal=4.4.0 | |
| 38 | - pygeos=0.9 | |
| 38 | - libpysal=4.5.1 | |
| 39 | - pygeos=0.10.2 | |
| 40 | - xyzservices=2021.9.1 | |
| 39 | 41 | - pip |
| 40 | 42 | - pip: |
| 41 | 43 | - sphinx-toggleprompt |
| 0 | 0 | /* colors */ |
| 1 | 1 | |
| 2 | h1 { | |
| 3 | color: #139C5A; | |
| 4 | } | |
| 5 | ||
| 6 | h2 { | |
| 7 | color: #333333; | |
| 8 | } | |
| 9 | ||
| 10 | .nav li.active>a, .navbar-nav>.active>.nav-link { | |
| 11 | color: #139C5A!important; | |
| 12 | } | |
| 13 | ||
| 14 | .toc-entry>.nav-link.active { | |
| 15 | border-left-color: #139C5A; | |
| 16 | color: #139C5A!important; | |
| 17 | } | |
| 18 | ||
| 19 | .nav li>a:hover { | |
| 20 | color: #333333!important; | |
| 2 | :root { | |
| 3 | --pst-color-primary: 19, 156, 90; | |
| 4 | --pst-color-active-navigation: 19, 156, 90; | |
| 5 | --pst-color-h2: var(--color-text-base); | |
| 21 | 6 | } |
| 22 | 7 | |
| 23 | 8 | /* buttons */ |
| 43 | 43 | imports when possible, and explicit relative imports for local |
| 44 | 44 | imports when necessary in tests. |
| 45 | 45 | |
| 46 | - GeoPandas supports Python 3.6+ only. The last version of GeoPandas | |
| 46 | - GeoPandas supports Python 3.7+ only. The last version of GeoPandas | |
| 47 | 47 | supporting Python 2 is 0.6. |
| 48 | 48 | |
| 49 | 49 | |
| 107 | 107 | the upstream (main project) *GeoPandas* repository. |
| 108 | 108 | |
| 109 | 109 | The testing suite will run automatically on GitHub Actions once your pull request is |
| 110 | submitted. The test suite will also autmatically run on your branch so you can | |
| 111 | check it prior to submitting the pull request. | |
| 110 | submitted. The test suite will also automatically run on your branch so you can | |
| 111 | check it prior to submitting the pull request. | |
| 112 | 112 | |
| 113 | 113 | Creating a branch |
| 114 | 114 | ~~~~~~~~~~~~~~~~~~ |
| 246 | 246 | 6) Updating the Documentation |
| 247 | 247 | ----------------------------- |
| 248 | 248 | |
| 249 | *GeoPandas* documentation resides in the ``doc`` folder. Changes to the docs are make by | |
| 250 | modifying the appropriate file in the `source` folder within ``doc``. *GeoPandas* docs use | |
| 249 | *GeoPandas* documentation resides in the ``doc`` folder. Changes to the docs are made by | |
| 250 | modifying the appropriate file in the ``source`` folder within ``doc``. *GeoPandas* docs use | |
| 251 | 251 | mixture of reStructuredText syntax for ``rst`` files, `which is explained here |
| 252 | 252 | <http://www.sphinx-doc.org/en/stable/rest.html#rst-primer>`_ and MyST syntax for ``md`` |
| 253 | 253 | files `explained here <https://myst-parser.readthedocs.io/en/latest/index.html>`_. |
| 256 | 256 | and examples are Jupyter notebooks converted to docs using `nbsphinx |
| 257 | 257 | <https://nbsphinx.readthedocs.io/>`_. Jupyter notebooks should be stored without the output. |
| 258 | 258 | |
| 259 | We highly encourage you to follow the `Google developer documentation style guide | |
| 260 | <https://developers.google.com/style/highlights>`_ when updating or creating new documentation. | |
| 261 | ||
| 259 | 262 | Once you have made your changes, you may try if they render correctly by |
| 260 | building the docs using sphinx. To do so, you can navigate to the `doc` folder | |
| 263 | building the docs using sphinx. To do so, you can navigate to the `doc` folder:: | |
| 264 | ||
| 265 | cd doc | |
| 266 | ||
| 261 | 267 | and type:: |
| 262 | 268 | |
| 263 | 269 | make html |
| 264 | 270 | |
| 265 | The resulting html pages will be located in ``doc/build/html``. In case of any errors, you | |
| 266 | can try to use ``make html`` within a new environment based on environment.yml | |
| 267 | specification in the ``doc`` folder. You may need to register Jupyter kernel as | |
| 271 | The resulting html pages will be located in ``doc/build/html``. | |
| 272 | ||
| 273 | In case of any errors, you can try to use ``make html`` within a new environment based on | |
| 274 | environment.yml specification in the ``doc`` folder. You may need to register Jupyter kernel as | |
| 268 | 275 | ``geopandas_docs``. Using conda:: |
| 269 | 276 | |
| 277 | cd doc | |
| 270 | 278 | conda env create -f environment.yml |
| 271 | 279 | conda activate geopandas_docs |
| 272 | 280 | python -m ipykernel install --user --name geopandas_docs |
| 273 | 281 | make html |
| 274 | 282 | |
| 275 | For minor updates, you can skip whole ``make html`` part as reStructuredText and MyST | |
| 283 | For minor updates, you can skip the ``make html`` part as reStructuredText and MyST | |
| 276 | 284 | syntax are usually quite straightforward. |
| 277 | 285 | |
| 278 | 286 | |
| 346 | 354 | Now you can commit your changes in your local repository:: |
| 347 | 355 | |
| 348 | 356 | git commit -m |
| 349 | ||
| 1 | 1 | |
| 2 | 2 | ## GeoPandas dependencies |
| 3 | 3 | |
| 4 | GeoPandas brings together the full capability of `pandas` and open-source geospatial | |
| 4 | GeoPandas brings together the full capability of `pandas` and the open-source geospatial | |
| 5 | 5 | tools `Shapely`, which brings manipulation and analysis of geometric objects backed by |
| 6 | 6 | [`GEOS`](https://trac.osgeo.org/geos) library, `Fiona`, allowing us to read and write |
| 7 | 7 | geographic data files using [`GDAL`](https://gdal.org), and `pyproj`, a library for |
| 8 | cartographic projections and coordinate transformations, which is a Python interface of | |
| 8 | cartographic projections and coordinate transformations, which is a Python interface to | |
| 9 | 9 | [`PROJ`](https://proj.org). |
| 10 | 10 | |
| 11 | 11 | Furthermore, GeoPandas has several optional dependencies as `rtree`, `pygeos`, |
| 39 | 39 | |
| 40 | 40 | #### [pyproj](https://github.com/pyproj4/pyproj) |
| 41 | 41 | `pyproj` is a Python interface to `PROJ` (cartographic projections and coordinate |
| 42 | transformations library). GeoPandas uses `pyproj.crs.CRS` object to keep track of a | |
| 42 | transformations library). GeoPandas uses a `pyproj.crs.CRS` object to keep track of the | |
| 43 | 43 | projection of each `GeoSeries` and its `Transformer` object to manage re-projections. |
| 44 | 44 | |
| 45 | 45 | ### Optional dependencies |
| 77 | 77 | |
| 78 | 78 | Various packages are built on top of GeoPandas addressing specific geospatial data |
| 79 | 79 | processing needs, analysis, and visualization. Below is an incomplete list (in no |
| 80 | particular order) of tools which form GeoPandas related Python ecosystem. | |
| 80 | particular order) of tools which form the GeoPandas-related Python ecosystem. | |
| 81 | 81 | |
| 82 | 82 | ### Spatial analysis and Machine Learning |
| 83 | 83 | |
| 104 | 104 | ##### [segregation](https://github.com/pysal/segregation) |
| 105 | 105 | `segregation` package calculates over 40 different segregation indices and provides a |
| 106 | 106 | suite of additional features for measurement, visualization, and hypothesis testing that |
| 107 | together represent the state-of-the-art in quantitative segregation analysis. | |
| 107 | together represent the state of the art in quantitative segregation analysis. | |
| 108 | 108 | |
| 109 | 109 | ##### [mgwr](https://github.com/pysal/mgwr) |
| 110 | 110 | `mgwr` provides scalable algorithms for estimation, inference, and prediction using |
| 111 | single- and multi-scale geographically-weighted regression models in a variety of | |
| 112 | generalized linear model frameworks, as well model diagnostics tools. | |
| 111 | single- and multi-scale geographically weighted regression models in a variety of | |
| 112 | generalized linear model frameworks, as well as model diagnostics tools. | |
| 113 | 113 | |
| 114 | 114 | ##### [tobler](https://github.com/pysal/tobler) |
| 115 | `tobler` provides functionality for for areal interpolation and dasymetric mapping. | |
| 115 | `tobler` provides functionality for areal interpolation and dasymetric mapping. | |
| 116 | 116 | `tobler` includes functionality for interpolating data using area-weighted approaches, |
| 117 | 117 | regression model-based approaches that leverage remotely-sensed raster data as auxiliary |
| 118 | 118 | information, and hybrid approaches. |
| 119 | ||
| 120 | 119 | |
| 121 | 120 | #### [movingpandas](https://github.com/anitagraser/movingpandas) |
| 122 | 121 | `MovingPandas` is a package for dealing with movement data. `MovingPandas` implements a |
| 162 | 161 | |
| 163 | 162 | ### Visualization |
| 164 | 163 | |
| 164 | #### [hvPlot](https://hvplot.holoviz.org/user_guide/Geographic_Data.html#Geopandas) | |
| 165 | `hvPlot` provides interactive Bokeh-based plotting for GeoPandas | |
| 166 | dataframes and series using the same API as the Matplotlib `.plot()` | |
| 167 | support that comes with GeoPandas. hvPlot makes it simple to pan and zoom into | |
| 168 | your plots, use widgets to explore multidimensional data, and render even the | |
| 169 | largest datasets in web browsers using [Datashader](https://datashader.org). | |
| 170 | ||
| 165 | 171 | #### [contextily](https://github.com/geopandas/contextily) |
| 166 | 172 | `contextily` is a small Python 3 (3.6 and above) package to retrieve tile maps from the |
| 167 | 173 | internet. It can add those tiles as basemap to `matplotlib` figures or write tile maps |
| 199 | 205 | `matplotlib`. |
| 200 | 206 | |
| 201 | 207 | #### [GeoViews](https://github.com/holoviz/geoviews) |
| 202 | `GeoViews` is a Python library that makes it easy to explore and visualize any data that | |
| 203 | includes geographic locations. It has particularly powerful support for multidimensional | |
| 204 | meteorological and oceanographic datasets, such as those used in weather, climate, and | |
| 205 | remote sensing research, but is useful for almost anything that you would want to plot | |
| 206 | on a map! | |
| 208 | `GeoViews` is a Python library that makes it easy to explore and | |
| 209 | visualize any data that includes geographic locations, with native | |
| 210 | support for GeoPandas dataframes and series objects. It has | |
| 211 | particularly powerful support for multidimensional meteorological and | |
| 212 | oceanographic datasets, such as those used in weather, climate, and | |
| 213 | remote sensing research, but is useful for almost anything that you | |
| 214 | would want to plot on a map! | |
| 207 | 215 | |
| 208 | 216 | #### [EarthPy](https://github.com/earthlab/earthpy) |
| 209 | 217 | `EarthPy` is a python package that makes it easier to plot and work with spatial raster |
| 228 | 236 | ### Geometry manipulation |
| 229 | 237 | |
| 230 | 238 | #### [TopoJSON](https://github.com/mattijn/topojson) |
| 231 | `Topojson` is a library that is capable of creating a topojson encoded format of merely | |
| 232 | any geographical object in Python. With topojson it is possible to reduce the size of | |
| 233 | your geographical data. Mostly by orders of magnitude. It is able to do so through: | |
| 234 | eliminating redundancy through computation of a topology; fixed-precision integer | |
| 235 | encoding of coordinates and simplification and quantization of arcs. | |
| 239 | `topojson` is a library for creating a TopoJSON encoding of nearly any | |
| 240 | geographical object in Python. With topojson it is possible to reduce the size of | |
| 241 | your geographical data, typically by orders of magnitude. It is able to do so through | |
| 242 | eliminating redundancy through computation of a topology, fixed-precision integer | |
| 243 | encoding of coordinates, and simplification and quantization of arcs. | |
| 236 | 244 | |
| 237 | 245 | #### [geocube](https://github.com/corteva/geocube) |
| 238 | 246 | Tool to convert geopandas vector data into rasterized `xarray` data. |
| 243 | 251 | `OSMnx` is a Python package that lets you download spatial data from OpenStreetMap and |
| 244 | 252 | model, project, visualize, and analyze real-world street networks. You can download and |
| 245 | 253 | model walkable, drivable, or bikeable urban networks with a single line of Python code |
| 246 | then easily analyze and visualize them. You can just as easily download and work with | |
| 254 | and then easily analyze and visualize them. You can just as easily download and work with | |
| 247 | 255 | other infrastructure types, amenities/points of interest, building footprints, elevation |
| 248 | 256 | data, street bearings/orientations, and speed/travel time. |
| 249 | 257 | |
| 266 | 274 | package is intended for exploratory data analysis and draws inspiration from |
| 267 | 275 | sqlalchemy-like interfaces and `acs.R`. With separate APIs for application developers |
| 268 | 276 | and folks who only want to get their data quickly & painlessly, `cenpy` should meet the |
| 269 | needs of most who aim to get US Census Data from Python. | |
| 277 | needs of most who aim to get US Census Data into Python. | |
| 270 | 278 | |
| 271 | 279 | ```{admonition} Expand this page |
| 272 | 280 | Do know a package which should be here? [Let us |
| 35 | 35 | "myst_parser", |
| 36 | 36 | "nbsphinx", |
| 37 | 37 | "numpydoc", |
| 38 | 'sphinx_toggleprompt', | |
| 39 | "matplotlib.sphinxext.plot_directive" | |
| 38 | "sphinx_toggleprompt", | |
| 39 | "matplotlib.sphinxext.plot_directive", | |
| 40 | 40 | ] |
| 41 | 41 | |
| 42 | 42 | # continue doc build and only print warnings/errors in examples |
| 53 | 53 | |
| 54 | 54 | |
| 55 | 55 | def setup(app): |
| 56 | app.add_stylesheet("custom.css") # may also be an URL | |
| 56 | app.add_css_file("custom.css") # may also be an URL | |
| 57 | 57 | |
| 58 | 58 | |
| 59 | 59 | # Add any paths that contain templates here, relative to this directory. |
| 64 | 64 | |
| 65 | 65 | nbsphinx_execute = "always" |
| 66 | 66 | nbsphinx_allow_errors = True |
| 67 | ||
| 68 | # connect docs in other projects | |
| 69 | intersphinx_mapping = {"pyproj": ("http://pyproj4.github.io/pyproj/stable/", None)} | |
| 67 | nbsphinx_kernel_name = "python3" | |
| 68 | ||
| 70 | 69 | # suppress matplotlib warning in examples |
| 71 | 70 | warnings.filterwarnings( |
| 72 | 71 | "ignore", |
| 330 | 329 | |
| 331 | 330 | __ https://github.com/geopandas/geopandas/blob/master/doc/source/{{ docname }} |
| 332 | 331 | """ |
| 332 | ||
| 333 | # --Options for sphinx extensions ----------------------------------------------- | |
| 334 | ||
| 335 | # connect docs in other projects | |
| 336 | intersphinx_mapping = { | |
| 337 | "cartopy": ( | |
| 338 | "https://scitools.org.uk/cartopy/docs/latest/", | |
| 339 | "https://scitools.org.uk/cartopy/docs/latest/objects.inv", | |
| 340 | ), | |
| 341 | "contextily": ( | |
| 342 | "https://contextily.readthedocs.io/en/stable/", | |
| 343 | "https://contextily.readthedocs.io/en/stable/objects.inv", | |
| 344 | ), | |
| 345 | "fiona": ( | |
| 346 | "https://fiona.readthedocs.io/en/stable/", | |
| 347 | "https://fiona.readthedocs.io/en/stable/objects.inv", | |
| 348 | ), | |
| 349 | "folium": ( | |
| 350 | "https://python-visualization.github.io/folium/", | |
| 351 | "https://python-visualization.github.io/folium/objects.inv", | |
| 352 | ), | |
| 353 | "geoplot": ( | |
| 354 | "https://residentmario.github.io/geoplot/index.html", | |
| 355 | "https://residentmario.github.io/geoplot/objects.inv", | |
| 356 | ), | |
| 357 | "geopy": ( | |
| 358 | "https://geopy.readthedocs.io/en/stable/", | |
| 359 | "https://geopy.readthedocs.io/en/stable/objects.inv", | |
| 360 | ), | |
| 361 | "libpysal": ( | |
| 362 | "https://pysal.org/libpysal/", | |
| 363 | "https://pysal.org/libpysal/objects.inv", | |
| 364 | ), | |
| 365 | "mapclassify": ( | |
| 366 | "https://pysal.org/mapclassify/", | |
| 367 | "https://pysal.org/mapclassify/objects.inv", | |
| 368 | ), | |
| 369 | "matplotlib": ( | |
| 370 | "https://matplotlib.org/stable/", | |
| 371 | "https://matplotlib.org/stable/objects.inv", | |
| 372 | ), | |
| 373 | "pandas": ( | |
| 374 | "https://pandas.pydata.org/pandas-docs/stable/", | |
| 375 | "https://pandas.pydata.org/pandas-docs/stable/objects.inv", | |
| 376 | ), | |
| 377 | "pyarrow": ("https://arrow.apache.org/docs/", None), | |
| 378 | "pyepsg": ( | |
| 379 | "https://pyepsg.readthedocs.io/en/stable/", | |
| 380 | "https://pyepsg.readthedocs.io/en/stable/objects.inv", | |
| 381 | ), | |
| 382 | "pygeos": ( | |
| 383 | "https://pygeos.readthedocs.io/en/latest/", | |
| 384 | "https://pygeos.readthedocs.io/en/latest/objects.inv", | |
| 385 | ), | |
| 386 | "pyproj": ( | |
| 387 | "https://pyproj4.github.io/pyproj/stable/", | |
| 388 | "https://pyproj4.github.io/pyproj/stable/objects.inv", | |
| 389 | ), | |
| 390 | "python": ( | |
| 391 | "https://docs.python.org/3", | |
| 392 | "https://docs.python.org/3/objects.inv", | |
| 393 | ), | |
| 394 | "rtree": ( | |
| 395 | "https://rtree.readthedocs.io/en/stable/", | |
| 396 | "https://rtree.readthedocs.io/en/stable/objects.inv", | |
| 397 | ), | |
| 398 | "rasterio": ( | |
| 399 | "https://rasterio.readthedocs.io/en/stable/", | |
| 400 | "https://rasterio.readthedocs.io/en/stable/objects.inv", | |
| 401 | ), | |
| 402 | "shapely": ( | |
| 403 | "https://shapely.readthedocs.io/en/stable/", | |
| 404 | "https://shapely.readthedocs.io/en/stable/objects.inv", | |
| 405 | ), | |
| 406 | "branca": ( | |
| 407 | "https://python-visualization.github.io/branca/", | |
| 408 | "https://python-visualization.github.io/branca/objects.inv", | |
| 409 | ), | |
| 410 | "xyzservices": ( | |
| 411 | "https://xyzservices.readthedocs.io/en/stable/", | |
| 412 | "https://xyzservices.readthedocs.io/en/stable/objects.inv", | |
| 413 | ), | |
| 414 | } | |
| 12 | 12 | |
| 13 | 13 | GeoDataFrame |
| 14 | 14 | |
| 15 | Reading and writing files | |
| 16 | ------------------------- | |
| 15 | Serialization / IO / conversion | |
| 16 | ------------------------------- | |
| 17 | 17 | |
| 18 | 18 | .. autosummary:: |
| 19 | 19 | :toctree: api/ |
| 26 | 26 | GeoDataFrame.to_parquet |
| 27 | 27 | GeoDataFrame.to_feather |
| 28 | 28 | GeoDataFrame.to_postgis |
| 29 | GeoDataFrame.to_wkb | |
| 30 | GeoDataFrame.to_wkt | |
| 29 | 31 | |
| 30 | 32 | Projection handling |
| 31 | 33 | ------------------- |
| 56 | 58 | GeoDataFrame.dissolve |
| 57 | 59 | GeoDataFrame.explode |
| 58 | 60 | |
| 61 | Spatial joins | |
| 62 | ------------- | |
| 63 | ||
| 64 | .. autosummary:: | |
| 65 | :toctree: api/ | |
| 66 | ||
| 67 | GeoDataFrame.sjoin | |
| 68 | GeoDataFrame.sjoin_nearest | |
| 69 | ||
| 70 | Overlay operations | |
| 71 | ------------------ | |
| 72 | ||
| 73 | .. autosummary:: | |
| 74 | :toctree: api/ | |
| 75 | ||
| 76 | GeoDataFrame.clip | |
| 77 | GeoDataFrame.overlay | |
| 78 | ||
| 59 | 79 | Plotting |
| 60 | 80 | -------- |
| 81 | ||
| 82 | .. autosummary:: | |
| 83 | :toctree: api/ | |
| 84 | ||
| 85 | GeoDataFrame.explore | |
| 86 | ||
| 61 | 87 | |
| 62 | 88 | .. autosummary:: |
| 63 | 89 | :toctree: api/ |
| 64 | 90 | :template: accessor_callable.rst |
| 65 | 91 | |
| 66 | 92 | GeoDataFrame.plot |
| 67 | ||
| 68 | 93 | |
| 69 | 94 | Spatial index |
| 70 | 95 | ------------- |
| 95 | 120 | All pandas ``DataFrame`` methods are also available, although they may |
| 96 | 121 | not operate in a meaningful way on the ``geometry`` column. All methods |
| 97 | 122 | listed in `GeoSeries <geoseries>`__ work directly on an active geometry column of GeoDataFrame. |
| 98 | ||
| 107 | 107 | GeoSeries.unary_union |
| 108 | 108 | GeoSeries.explode |
| 109 | 109 | |
| 110 | Reading and writing files | |
| 111 | ------------------------- | |
| 110 | Serialization / IO / conversion | |
| 111 | ------------------------------- | |
| 112 | 112 | |
| 113 | 113 | .. autosummary:: |
| 114 | 114 | :toctree: api/ |
| 115 | 115 | |
| 116 | 116 | GeoSeries.from_file |
| 117 | GeoSeries.from_wkb | |
| 118 | GeoSeries.from_wkt | |
| 119 | GeoSeries.from_xy | |
| 117 | 120 | GeoSeries.to_file |
| 118 | 121 | GeoSeries.to_json |
| 122 | GeoSeries.to_wkb | |
| 123 | GeoSeries.to_wkt | |
| 119 | 124 | |
| 120 | 125 | Projection handling |
| 121 | 126 | ------------------- |
| 138 | 143 | GeoSeries.isna |
| 139 | 144 | GeoSeries.notna |
| 140 | 145 | |
| 146 | Overlay operations | |
| 147 | ------------------ | |
| 148 | ||
| 149 | .. autosummary:: | |
| 150 | :toctree: api/ | |
| 151 | ||
| 152 | GeoSeries.clip | |
| 153 | ||
| 141 | 154 | Plotting |
| 142 | 155 | -------- |
| 143 | 156 | |
| 145 | 158 | :toctree: api/ |
| 146 | 159 | |
| 147 | 160 | GeoSeries.plot |
| 161 | GeoSeries.explore | |
| 148 | 162 | |
| 149 | 163 | |
| 150 | 164 | Spatial index |
| 29 | 29 | |
| 30 | 30 | intersection |
| 31 | 31 | is_empty |
| 32 | nearest | |
| 32 | 33 | query |
| 33 | 34 | query_bulk |
| 34 | 35 | size |
| 41 | 42 | (``geopandas.sindex.RTreeIndex``) offers the full capability of |
| 42 | 43 | ``rtree.index.Index`` - see the full API in the `rtree documentation`_. |
| 43 | 44 | |
| 45 | Similarly, the ``pygeos``-based spatial index | |
| 46 | (``geopandas.sindex.PyGEOSSTRTreeIndex``) offers the full capability of | |
| 47 | ``pygeos.STRtree``, including nearest-neighbor queries. | |
| 48 | See the full API in the `PyGEOS STRTree documentation`_. | |
| 49 | ||
| 44 | 50 | .. _rtree documentation: https://rtree.readthedocs.io/en/stable/class.html |
| 51 | .. _PyGEOS STRTree documentation: https://pygeos.readthedocs.io/en/latest/strtree.html | |
| 11 | 11 | |
| 12 | 12 | Spatial data are often more granular than we need. For example, we might have data on sub-national units, but we're actually interested in studying patterns at the level of countries. |
| 13 | 13 | |
| 14 | In a non-spatial setting, when all we need are summary statistics of the data, we aggregate our data using the ``groupby`` function. But for spatial data, we sometimes also need to aggregate geometric features. In the *geopandas* library, we can aggregate geometric features using the ``dissolve`` function. | |
| 14 | In a non-spatial setting, when all we need are summary statistics of the data, we aggregate our data using the :meth:`~pandas.DataFrame.groupby` function. But for spatial data, we sometimes also need to aggregate geometric features. In the *geopandas* library, we can aggregate geometric features using the :meth:`~geopandas.GeoDataFrame.dissolve` function. | |
| 15 | 15 | |
| 16 | ``dissolve`` can be thought of as doing three things: (a) it dissolves all the geometries within a given group together into a single geometric feature (using the ``unary_union`` method), and (b) it aggregates all the rows of data in a group using ``groupby.aggregate()``, and (c) it combines those two results. | |
| 16 | :meth:`~geopandas.GeoDataFrame.dissolve` can be thought of as doing three things: | |
| 17 | 17 | |
| 18 | ``dissolve`` Example | |
| 19 | ~~~~~~~~~~~~~~~~~~~~~ | |
| 18 | (a) it dissolves all the geometries within a given group together into a single geometric feature (using the :attr:`~geopandas.GeoSeries.unary_union` method), and | |
| 19 | (b) it aggregates all the rows of data in a group using :ref:`groupby.aggregate <groupby.aggregate>`, and | |
| 20 | (c) it combines those two results. | |
| 21 | ||
| 22 | :meth:`~geopandas.GeoDataFrame.dissolve` Example | |
| 23 | ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~ | |
| 20 | 24 | |
| 21 | 25 | Suppose we are interested in studying continents, but we only have country-level data like the country dataset included in *geopandas*. We can easily convert this to a continent-level dataset. |
| 22 | 26 | |
| 23 | 27 | |
| 24 | First, let's look at the most simple case where we just want continent shapes and names. By default, ``dissolve`` will pass ``'first'`` to ``groupby.aggregate``. | |
| 28 | First, let's look at the most simple case where we just want continent shapes and names. By default, :meth:`~geopandas.GeoDataFrame.dissolve` will pass ``'first'`` to :ref:`groupby.aggregate <groupby.aggregate>`. | |
| 25 | 29 | |
| 26 | 30 | .. ipython:: python |
| 27 | 31 | |
| 34 | 38 | |
| 35 | 39 | continents.head() |
| 36 | 40 | |
| 37 | If we are interested in aggregate populations, however, we can pass different functions to the ``dissolve`` method to aggregate populations using the ``aggfunc =`` argument: | |
| 41 | If we are interested in aggregate populations, however, we can pass different functions to the :meth:`~geopandas.GeoDataFrame.dissolve` method to aggregate populations using the ``aggfunc =`` argument: | |
| 38 | 42 | |
| 39 | 43 | .. ipython:: python |
| 40 | 44 | |
| 61 | 65 | ~~~~~~~~~~~~~~~~~~ |
| 62 | 66 | |
| 63 | 67 | The ``aggfunc =`` argument defaults to 'first' which means that the first row of attributes values found in the dissolve routine will be assigned to the resultant dissolved geodataframe. |
| 64 | However it also accepts other summary statistic options as allowed by ``pandas.groupby()`` including: | |
| 68 | However it also accepts other summary statistic options as allowed by :meth:`pandas.groupby <pandas.DataFrame.groupby>` including: | |
| 65 | 69 | |
| 66 | 70 | * 'first' |
| 67 | 71 | * 'last' |
| 13 | 13 | ========================================= |
| 14 | 14 | |
| 15 | 15 | GeoPandas implements two main data structures, a :class:`GeoSeries` and a |
| 16 | :class:`GeoDataFrame`. These are subclasses of pandas ``Series`` and | |
| 17 | ``DataFrame``, respectively. | |
| 16 | :class:`GeoDataFrame`. These are subclasses of :class:`pandas.Series` and | |
| 17 | :class:`pandas.DataFrame`, respectively. | |
| 18 | 18 | |
| 19 | 19 | GeoSeries |
| 20 | 20 | --------- |
| 44 | 44 | by matching indices. Binary operations can also be applied to a |
| 45 | 45 | single geometry, in which case the operation is carried out for each |
| 46 | 46 | element of the series with that geometry. In either case, a |
| 47 | ``Series`` or a :class:`GeoSeries` will be returned, as appropriate. | |
| 47 | :class:`~pandas.Series` or a :class:`GeoSeries` will be returned, as appropriate. | |
| 48 | 48 | |
| 49 | 49 | A short summary of a few attributes and methods for GeoSeries is |
| 50 | 50 | presented here, and a full list can be found in the :doc:`all attributes and methods page <../reference/geoseries>`. |
| 63 | 63 | Basic Methods |
| 64 | 64 | ^^^^^^^^^^^^^^ |
| 65 | 65 | |
| 66 | * :meth:`~GeoSeries.distance`: returns ``Series`` with minimum distance from each entry to ``other`` | |
| 66 | * :meth:`~GeoSeries.distance`: returns :class:`~pandas.Series` with minimum distance from each entry to ``other`` | |
| 67 | 67 | * :attr:`~GeoSeries.centroid`: returns :class:`GeoSeries` of centroids |
| 68 | 68 | * :meth:`~GeoSeries.representative_point`: returns :class:`GeoSeries` of points that are guaranteed to be within each geometry. It does **NOT** return centroids. |
| 69 | 69 | * :meth:`~GeoSeries.to_crs`: change coordinate reference system. See :doc:`projections <projections>` |
| 116 | 116 | Now, we create centroids and make it the geometry: |
| 117 | 117 | |
| 118 | 118 | .. ipython:: python |
| 119 | :okwarning: | |
| 119 | 120 | |
| 120 | 121 | world['centroid_column'] = world.centroid |
| 121 | 122 | world = world.set_geometry('centroid_column') |
| 128 | 129 | |
| 129 | 130 | gdf = gdf.rename(columns={'old_name': 'new_name'}).set_geometry('new_name') |
| 130 | 131 | |
| 131 | **Note 2:** Somewhat confusingly, by default when you use the ``read_file`` command, the column containing spatial objects from the file is named "geometry" by default, and will be set as the active geometry column. However, despite using the same term for the name of the column and the name of the special attribute that keeps track of the active column, they are distinct. You can easily shift the active geometry column to a different :class:`GeoSeries` with the :meth:`~GeoDataFrame.set_geometry` command. Further, ``gdf.geometry`` will always return the active geometry column, *not* the column named ``geometry``. If you wish to call a column named "geometry", and a different column is the active geometry column, use ``gdf['geometry']``, not ``gdf.geometry``. | |
| 132 | **Note 2:** Somewhat confusingly, by default when you use the :func:`~geopandas.read_file` command, the column containing spatial objects from the file is named "geometry" by default, and will be set as the active geometry column. However, despite using the same term for the name of the column and the name of the special attribute that keeps track of the active column, they are distinct. You can easily shift the active geometry column to a different :class:`GeoSeries` with the :meth:`~GeoDataFrame.set_geometry` command. Further, ``gdf.geometry`` will always return the active geometry column, *not* the column named ``geometry``. If you wish to call a column named "geometry", and a different column is the active geometry column, use ``gdf['geometry']``, not ``gdf.geometry``. | |
| 132 | 133 | |
| 133 | 134 | Attributes and Methods |
| 134 | 135 | ~~~~~~~~~~~~~~~~~~~~~~ |
| 135 | 136 | |
| 136 | 137 | Any of the attributes calls or methods described for a :class:`GeoSeries` will work on a :class:`GeoDataFrame` -- effectively, they are just applied to the "geometry" :class:`GeoSeries`. |
| 137 | 138 | |
| 138 | However, ``GeoDataFrames`` also have a few extra methods for input and output which are described on the :doc:`Input and Output <io>` page and for geocoding with are described in :doc:`Geocoding <geocoding>`. | |
| 139 | However, :class:`GeoDataFrames <GeoDataFrame>` also have a few extra methods for input and output which are described on the :doc:`Input and Output <io>` page and for geocoding with are described in :doc:`Geocoding <geocoding>`. | |
| 139 | 140 | |
| 140 | 141 | |
| 141 | 142 | .. ipython:: python |
| 156 | 157 | geopandas.options |
| 157 | 158 | |
| 158 | 159 | The ``geopandas.options.display_precision`` option can control the number of |
| 159 | decimals to show in the display of coordinates in the geometry column. | |
| 160 | decimals to show in the display of coordinates in the geometry column. | |
| 160 | 161 | In the ``world`` example of above, the default is to show 5 decimals for |
| 161 | 162 | geographic coordinates: |
| 162 | 163 | |
| 31 | 31 | boro_locations.plot(ax=ax, color="red"); |
| 32 | 32 | |
| 33 | 33 | |
| 34 | By default, the ``geocode`` function uses the | |
| 35 | `GeoCode.Farm geocoding API <https://geocode.farm/>`__ with a rate limitation | |
| 36 | applied. But a different geocoding service can be specified with the | |
| 34 | By default, the :func:`~geopandas.tools.geocode` function uses the | |
| 35 | `Photon geocoding API <https://photon.komoot.io>`__. | |
| 36 | But a different geocoding service can be specified with the | |
| 37 | 37 | ``provider`` keyword. |
| 38 | 38 | |
| 39 | 39 | The argument to ``provider`` can either be a string referencing geocoding |
| 40 | 40 | services, such as ``'google'``, ``'bing'``, ``'yahoo'``, and |
| 41 | ``'openmapquest'``, or an instance of a ``Geocoder`` from ``geopy``. See | |
| 41 | ``'openmapquest'``, or an instance of a :mod:`Geocoder <geopy.geocoders>` from :mod:`geopy`. See | |
| 42 | 42 | ``geopy.geocoders.SERVICE_TO_GEOCODER`` for the full list. |
| 43 | 43 | For many providers, parameters such as API keys need to be passed as |
| 44 | ``**kwargs`` in the ``geocode`` call. | |
| 44 | ``**kwargs`` in the :func:`~geopandas.tools.geocode` call. | |
| 45 | 45 | |
| 46 | 46 | For example, to use the OpenStreetMap Nominatim geocoder, you need to specify |
| 47 | 47 | a user agent: |
| 53 | 53 | .. attention:: |
| 54 | 54 | |
| 55 | 55 | Please consult the Terms of Service for the chosen provider. The example |
| 56 | above uses ``'geocodefarm'`` (the default), for which free users are | |
| 57 | limited to 250 calls per day and 4 requests per second | |
| 58 | (`geocodefarm ToS <https://geocode.farm/geocoding/free-api-documentation/>`_). | |
| 56 | above uses ``'photon'`` (the default), which expects fair usage | |
| 57 | - extensive usage will be throttled. | |
| 58 | (`Photon's Terms of Use <https://photon.komoot.io>`_). | |
| 11 | 11 | |
| 12 | 12 | .. method:: GeoSeries.buffer(distance, resolution=16) |
| 13 | 13 | |
| 14 | Returns a ``GeoSeries`` of geometries representing all points within a given `distance` | |
| 14 | Returns a :class:`~geopandas.GeoSeries` of geometries representing all points within a given `distance` | |
| 15 | 15 | of each geometric object. |
| 16 | 16 | |
| 17 | 17 | .. attribute:: GeoSeries.boundary |
| 18 | 18 | |
| 19 | Returns a ``GeoSeries`` of lower dimensional objects representing | |
| 19 | Returns a :class:`~geopandas.GeoSeries` of lower dimensional objects representing | |
| 20 | 20 | each geometries's set-theoretic `boundary`. |
| 21 | 21 | |
| 22 | 22 | .. attribute:: GeoSeries.centroid |
| 23 | 23 | |
| 24 | Returns a ``GeoSeries`` of points for each geometric centroid. | |
| 24 | Returns a :class:`~geopandas.GeoSeries` of points for each geometric centroid. | |
| 25 | 25 | |
| 26 | 26 | .. attribute:: GeoSeries.convex_hull |
| 27 | 27 | |
| 28 | Returns a ``GeoSeries`` of geometries representing the smallest | |
| 28 | Returns a :class:`~geopandas.GeoSeries` of geometries representing the smallest | |
| 29 | 29 | convex `Polygon` containing all the points in each object unless the |
| 30 | 30 | number of points in the object is less than three. For two points, |
| 31 | 31 | the convex hull collapses to a `LineString`; for 1, a `Point`. |
| 32 | 32 | |
| 33 | 33 | .. attribute:: GeoSeries.envelope |
| 34 | 34 | |
| 35 | Returns a ``GeoSeries`` of geometries representing the point or | |
| 35 | Returns a :class:`~geopandas.GeoSeries` of geometries representing the point or | |
| 36 | 36 | smallest rectangular polygon (with sides parallel to the coordinate |
| 37 | 37 | axes) that contains each object. |
| 38 | 38 | |
| 39 | 39 | .. method:: GeoSeries.simplify(tolerance, preserve_topology=True) |
| 40 | 40 | |
| 41 | Returns a ``GeoSeries`` containing a simplified representation of | |
| 41 | Returns a :class:`~geopandas.GeoSeries` containing a simplified representation of | |
| 42 | 42 | each object. |
| 43 | 43 | |
| 44 | 44 | .. attribute:: GeoSeries.unary_union |
| 45 | 45 | |
| 46 | Return a geometry containing the union of all geometries in the ``GeoSeries``. | |
| 46 | Return a geometry containing the union of all geometries in the :class:`~geopandas.GeoSeries`. | |
| 47 | 47 | |
| 48 | 48 | |
| 49 | 49 | Affine transformations |
| 51 | 51 | |
| 52 | 52 | .. method:: GeoSeries.affine_transform(self, matrix) |
| 53 | 53 | |
| 54 | Transform the geometries of the GeoSeries using an affine transformation matrix | |
| 54 | Transform the geometries of the :class:`~geopandas.GeoSeries` using an affine transformation matrix | |
| 55 | 55 | |
| 56 | 56 | .. method:: GeoSeries.rotate(self, angle, origin='center', use_radians=False) |
| 57 | 57 | |
| 58 | Rotate the coordinates of the GeoSeries. | |
| 58 | Rotate the coordinates of the :class:`~geopandas.GeoSeries`. | |
| 59 | 59 | |
| 60 | 60 | .. method:: GeoSeries.scale(self, xfact=1.0, yfact=1.0, zfact=1.0, origin='center') |
| 61 | 61 | |
| 62 | Scale the geometries of the GeoSeries along each (x, y, z) dimensio. | |
| 62 | Scale the geometries of the :class:`~geopandas.GeoSeries` along each (x, y, z) dimensio. | |
| 63 | 63 | |
| 64 | 64 | .. method:: GeoSeries.skew(self, angle, origin='center', use_radians=False) |
| 65 | 65 | |
| 66 | Shear/Skew the geometries of the GeoSeries by angles along x and y dimensions. | |
| 66 | Shear/Skew the geometries of the :class:`~geopandas.GeoSeries` by angles along x and y dimensions. | |
| 67 | 67 | |
| 68 | 68 | .. method:: GeoSeries.translate(self, xoff=0.0, yoff=0.0, zoff=0.0) |
| 69 | 69 | |
| 70 | Shift the coordinates of the GeoSeries. | |
| 70 | Shift the coordinates of the :class:`~geopandas.GeoSeries`. | |
| 71 | 71 | |
| 72 | 72 | |
| 73 | 73 | |
| 91 | 91 | |
| 92 | 92 | .. image:: ../../_static/test.png |
| 93 | 93 | |
| 94 | Some geographic operations return normal pandas object. The ``area`` property of a ``GeoSeries`` will return a ``pandas.Series`` containing the area of each item in the ``GeoSeries``: | |
| 94 | Some geographic operations return normal pandas object. The :attr:`~geopandas.GeoSeries.area` property of a :class:`~geopandas.GeoSeries` will return a :class:`pandas.Series` containing the area of each item in the :class:`~geopandas.GeoSeries`: | |
| 95 | 95 | |
| 96 | 96 | .. sourcecode:: python |
| 97 | 97 | |
| 160 | 160 | .. image:: ../../_static/nyc_hull.png |
| 161 | 161 | |
| 162 | 162 | To demonstrate a more complex operation, we'll generate a |
| 163 | ``GeoSeries`` containing 2000 random points: | |
| 163 | :class:`~geopandas.GeoSeries` containing 2000 random points: | |
| 164 | 164 | |
| 165 | 165 | .. sourcecode:: python |
| 166 | 166 | |
| 177 | 177 | |
| 178 | 178 | >>> circles = pts.buffer(2000) |
| 179 | 179 | |
| 180 | We can collapse these circles into a single shapely MultiPolygon | |
| 180 | We can collapse these circles into a single :class:`MultiPolygon` | |
| 181 | 181 | geometry with |
| 182 | 182 | |
| 183 | 183 | .. sourcecode:: python |
| 202 | 202 | .. image:: ../../_static/boros_with_holes.png |
| 203 | 203 | |
| 204 | 204 | Note that this can be simplified a bit, since ``geometry`` is |
| 205 | available as an attribute on a ``GeoDataFrame``, and the | |
| 206 | ``intersection`` and ``difference`` methods are implemented with the | |
| 205 | available as an attribute on a :class:`~geopandas.GeoDataFrame`, and the | |
| 206 | :meth:`~geopandas.GeoSeries.intersection` and :meth:`~geopandas.GeoSeries.difference` methods are implemented with the | |
| 207 | 207 | "&" and "-" operators, respectively. For example, the latter could |
| 208 | 208 | have been expressed simply as ``boros.geometry - mp``. |
| 209 | 209 | |
| 8 | 8 | Indexing and Selecting Data |
| 9 | 9 | =========================== |
| 10 | 10 | |
| 11 | GeoPandas inherits the standard ``pandas`` methods for indexing/selecting data. This includes label based indexing with ``.loc`` and integer position based indexing with ``.iloc``, which apply to both ``GeoSeries`` and ``GeoDataFrame`` objects. For more information on indexing/selecting, see the pandas_ documentation. | |
| 11 | GeoPandas inherits the standard pandas_ methods for indexing/selecting data. This includes label based indexing with :attr:`~pandas.DataFrame.loc` and integer position based indexing with :attr:`~pandas.DataFrame.iloc`, which apply to both :class:`GeoSeries` and :class:`GeoDataFrame` objects. For more information on indexing/selecting, see the pandas_ documentation. | |
| 12 | 12 | |
| 13 | 13 | .. _pandas: http://pandas.pydata.org/pandas-docs/stable/indexing.html |
| 14 | 14 | |
| 15 | In addition to the standard ``pandas`` methods, GeoPandas also provides | |
| 16 | coordinate based indexing with the ``cx`` indexer, which slices using a bounding | |
| 17 | box. Geometries in the ``GeoSeries`` or ``GeoDataFrame`` that intersect the | |
| 15 | In addition to the standard pandas_ methods, GeoPandas also provides | |
| 16 | coordinate based indexing with the :attr:`~GeoDataFrame.cx` indexer, which slices using a bounding | |
| 17 | box. Geometries in the :class:`GeoSeries` or :class:`GeoDataFrame` that intersect the | |
| 18 | 18 | bounding box will be returned. |
| 19 | 19 | |
| 20 | 20 | Using the ``world`` dataset, we can use this functionality to quickly select all |
| 26 | 26 | southern_world = world.cx[:, :0] |
| 27 | 27 | @savefig world_southern.png |
| 28 | 28 | southern_world.plot(figsize=(10, 3)); |
| 29 | ||
| 0 | { | |
| 1 | "cells": [ | |
| 2 | { | |
| 3 | "cell_type": "markdown", | |
| 4 | "source": [ | |
| 5 | "# Interactive mapping\n", | |
| 6 | "\n", | |
| 7 | "Alongside static plots, `geopandas` can create interactive maps based on the [folium](https://python-visualization.github.io/folium/) library.\n", | |
| 8 | "\n", | |
| 9 | "Creating maps for interactive exploration mirrors the API of [static plots](../reference/api/geopandas.GeoDataFrame.plot.html) in an [explore()](../reference/api/geopandas.GeoDataFrame.explore.html) method of a GeoSeries or GeoDataFrame.\n", | |
| 10 | "\n", | |
| 11 | "Loading some example data:" | |
| 12 | ], | |
| 13 | "metadata": {} | |
| 14 | }, | |
| 15 | { | |
| 16 | "cell_type": "code", | |
| 17 | "execution_count": null, | |
| 18 | "source": [ | |
| 19 | "import geopandas\n", | |
| 20 | "\n", | |
| 21 | "nybb = geopandas.read_file(geopandas.datasets.get_path('nybb'))\n", | |
| 22 | "world = geopandas.read_file(geopandas.datasets.get_path('naturalearth_lowres'))\n", | |
| 23 | "cities = geopandas.read_file(geopandas.datasets.get_path('naturalearth_cities'))" | |
| 24 | ], | |
| 25 | "outputs": [], | |
| 26 | "metadata": {} | |
| 27 | }, | |
| 28 | { | |
| 29 | "cell_type": "markdown", | |
| 30 | "source": [ | |
| 31 | "The simplest option is to use `GeoDataFrame.explore()`:" | |
| 32 | ], | |
| 33 | "metadata": {} | |
| 34 | }, | |
| 35 | { | |
| 36 | "cell_type": "code", | |
| 37 | "execution_count": null, | |
| 38 | "source": [ | |
| 39 | "nybb.explore()" | |
| 40 | ], | |
| 41 | "outputs": [], | |
| 42 | "metadata": {} | |
| 43 | }, | |
| 44 | { | |
| 45 | "cell_type": "markdown", | |
| 46 | "source": [ | |
| 47 | "Interactive plotting offers largely the same customisation as static one plus some features on top of that. Check the code below which plots a customised choropleth map. You can use `\"BoroName\"` column with NY boroughs names as an input of the choropleth, show (only) its name in the tooltip on hover but show all values on click. You can also pass custom background tiles (either a name supported by folium, a name recognized by `xyzservices.providers.query_name()`, XYZ URL or `xyzservices.TileProvider` object), specify colormap (all supported by `matplotlib`) and specify black outline." | |
| 48 | ], | |
| 49 | "metadata": {} | |
| 50 | }, | |
| 51 | { | |
| 52 | "cell_type": "code", | |
| 53 | "execution_count": null, | |
| 54 | "source": [ | |
| 55 | "nybb.explore( \n", | |
| 56 | " column=\"BoroName\", # make choropleth based on \"BoroName\" column\n", | |
| 57 | " tooltip=\"BoroName\", # show \"BoroName\" value in tooltip (on hover)\n", | |
| 58 | " popup=True, # show all values in popup (on click)\n", | |
| 59 | " tiles=\"CartoDB positron\", # use \"CartoDB positron\" tiles\n", | |
| 60 | " cmap=\"Set1\", # use \"Set1\" matplotlib colormap\n", | |
| 61 | " style_kwds=dict(color=\"black\") # use black outline\n", | |
| 62 | " )" | |
| 63 | ], | |
| 64 | "outputs": [], | |
| 65 | "metadata": {} | |
| 66 | }, | |
| 67 | { | |
| 68 | "cell_type": "markdown", | |
| 69 | "source": [ | |
| 70 | "The `explore()` method returns a `folium.Map` object, which can also be passed directly (as you do with `ax` in `plot()`). You can then use folium functionality directly on the resulting map. In the example below, you can plot two GeoDataFrames on the same map and add layer control using folium. You can also add additional tiles allowing you to change the background directly in the map." | |
| 71 | ], | |
| 72 | "metadata": {} | |
| 73 | }, | |
| 74 | { | |
| 75 | "cell_type": "code", | |
| 76 | "execution_count": null, | |
| 77 | "source": [ | |
| 78 | "import folium\n", | |
| 79 | "\n", | |
| 80 | "m = world.explore(\n", | |
| 81 | " column=\"pop_est\", # make choropleth based on \"BoroName\" column\n", | |
| 82 | " scheme=\"naturalbreaks\", # use mapclassify's natural breaks scheme\n", | |
| 83 | " legend=True, # show legend\n", | |
| 84 | " k=10, # use 10 bins\n", | |
| 85 | " legend_kwds=dict(colorbar=False), # do not use colorbar\n", | |
| 86 | " name=\"countries\" # name of the layer in the map\n", | |
| 87 | ")\n", | |
| 88 | "\n", | |
| 89 | "cities.explore(\n", | |
| 90 | " m=m, # pass the map object\n", | |
| 91 | " color=\"red\", # use red color on all points\n", | |
| 92 | " marker_kwds=dict(radius=10, fill=True), # make marker radius 10px with fill\n", | |
| 93 | " tooltip=\"name\", # show \"name\" column in the tooltip\n", | |
| 94 | " tooltip_kwds=dict(labels=False), # do not show column label in the tooltip\n", | |
| 95 | " name=\"cities\" # name of the layer in the map\n", | |
| 96 | ")\n", | |
| 97 | "\n", | |
| 98 | "folium.TileLayer('Stamen Toner', control=True).add_to(m) # use folium to add alternative tiles\n", | |
| 99 | "folium.LayerControl().add_to(m) # use folium to add layer control\n", | |
| 100 | "\n", | |
| 101 | "m # show map" | |
| 102 | ], | |
| 103 | "outputs": [], | |
| 104 | "metadata": {} | |
| 105 | } | |
| 106 | ], | |
| 107 | "metadata": { | |
| 108 | "kernelspec": { | |
| 109 | "display_name": "Python 3", | |
| 110 | "language": "python", | |
| 111 | "name": "python3" | |
| 112 | }, | |
| 113 | "language_info": { | |
| 114 | "codemirror_mode": { | |
| 115 | "name": "ipython", | |
| 116 | "version": 3 | |
| 117 | }, | |
| 118 | "file_extension": ".py", | |
| 119 | "mimetype": "text/x-python", | |
| 120 | "name": "python", | |
| 121 | "nbconvert_exporter": "python", | |
| 122 | "pygments_lexer": "ipython3", | |
| 123 | "version": "3.9.2" | |
| 124 | } | |
| 125 | }, | |
| 126 | "nbformat": 4, | |
| 127 | "nbformat_minor": 5 | |
| 128 | }⏎ |
| 17 | 17 | transformations. |
| 18 | 18 | |
| 19 | 19 | Any arguments passed to :func:`geopandas.read_file` after the file name will be |
| 20 | passed directly to ``fiona.open``, which does the actual data importation. In | |
| 20 | passed directly to :func:`fiona.open`, which does the actual data importation. In | |
| 21 | 21 | general, :func:`geopandas.read_file` is pretty smart and should do what you want |
| 22 | 22 | without extra arguments, but for more help, type:: |
| 23 | 23 | |
| 29 | 29 | |
| 30 | 30 | countries_gdf = geopandas.read_file("package.gpkg", layer='countries') |
| 31 | 31 | |
| 32 | Where supported in ``fiona``, *geopandas* can also load resources directly from | |
| 32 | Where supported in :mod:`fiona`, *geopandas* can also load resources directly from | |
| 33 | 33 | a web URL, for example for GeoJSON files from `geojson.xyz <http://geojson.xyz/>`_:: |
| 34 | 34 | |
| 35 | 35 | url = "http://d2ad6b4ur7yvpq.cloudfront.net/naturalearth-3.3.0/ne_110m_land.geojson" |
| 49 | 49 | |
| 50 | 50 | zipfile = "zip:///Users/name/Downloads/gadm36_AFG_shp.zip!data/gadm36_AFG_1.shp" |
| 51 | 51 | |
| 52 | It is also possible to read any file-like objects with a ``read()`` method, such | |
| 53 | as a file handler (e.g. via built-in ``open`` function) or ``StringIO``:: | |
| 52 | It is also possible to read any file-like objects with a :func:`os.read` method, such | |
| 53 | as a file handler (e.g. via built-in :func:`open` function) or :class:`~io.StringIO`:: | |
| 54 | 54 | |
| 55 | 55 | filename = "test.geojson" |
| 56 | 56 | file = open(filename) |
| 196 | 196 | Writing to PostGIS:: |
| 197 | 197 | |
| 198 | 198 | from sqlalchemy import create_engine |
| 199 | db_connection_url = "postgres://myusername:mypassword@myhost:5432/mydatabase"; | |
| 199 | db_connection_url = "postgresql://myusername:mypassword@myhost:5432/mydatabase"; | |
| 200 | 200 | engine = create_engine(db_connection_url) |
| 201 | 201 | countries_gdf.to_postgis("countries_table", con=engine) |
| 202 | 202 | |
| 14 | 14 | ========================================= |
| 15 | 15 | |
| 16 | 16 | |
| 17 | *geopandas* provides a high-level interface to the ``matplotlib`` library for making maps. Mapping shapes is as easy as using the ``plot()`` method on a ``GeoSeries`` or ``GeoDataFrame``. | |
| 17 | *geopandas* provides a high-level interface to the matplotlib_ library for making maps. Mapping shapes is as easy as using the :meth:`~GeoDataFrame.plot()` method on a :class:`GeoSeries` or :class:`GeoDataFrame`. | |
| 18 | ||
| 19 | .. _matplotlib: https://matplotlib.org/stable/ | |
| 18 | 20 | |
| 19 | 21 | Loading some example data: |
| 20 | 22 | |
| 34 | 36 | @savefig world_randomcolors.png |
| 35 | 37 | world.plot(); |
| 36 | 38 | |
| 37 | Note that in general, any options one can pass to `pyplot <http://matplotlib.org/api/pyplot_api.html>`_ in ``matplotlib`` (or `style options that work for lines <http://matplotlib.org/api/lines_api.html>`_) can be passed to the ``plot()`` method. | |
| 39 | Note that in general, any options one can pass to `pyplot <http://matplotlib.org/api/pyplot_api.html>`_ in matplotlib_ (or `style options that work for lines <http://matplotlib.org/api/lines_api.html>`_) can be passed to the :meth:`~GeoDataFrame.plot` method. | |
| 38 | 40 | |
| 39 | 41 | |
| 40 | 42 | Choropleth Maps |
| 43 | 45 | *geopandas* makes it easy to create Choropleth maps (maps where the color of each shape is based on the value of an associated variable). Simply use the plot command with the ``column`` argument set to the column whose values you want used to assign colors. |
| 44 | 46 | |
| 45 | 47 | .. ipython:: python |
| 46 | ||
| 47 | # Plot by GDP per capta | |
| 48 | :okwarning: | |
| 49 | ||
| 50 | # Plot by GDP per capita | |
| 48 | 51 | world = world[(world.pop_est>0) & (world.name!="Antarctica")] |
| 49 | 52 | world['gdp_per_cap'] = world.gdp_md_est / world.pop_est |
| 50 | 53 | @savefig world_gdp_per_cap.png |
| 64 | 67 | @savefig world_pop_est.png |
| 65 | 68 | world.plot(column='pop_est', ax=ax, legend=True) |
| 66 | 69 | |
| 67 | However, the default appearance of the legend and plot axes may not be desirable. One can define the plot axes (with ``ax``) and the legend axes (with ``cax``) and then pass those in to the ``plot`` call. The following example uses ``mpl_toolkits`` to vertically align the plot axes and the legend axes: | |
| 70 | However, the default appearance of the legend and plot axes may not be desirable. One can define the plot axes (with ``ax``) and the legend axes (with ``cax``) and then pass those in to the :meth:`~GeoDataFrame.plot` call. The following example uses ``mpl_toolkits`` to vertically align the plot axes and the legend axes: | |
| 68 | 71 | |
| 69 | 72 | .. ipython:: python |
| 70 | 73 | |
| 95 | 98 | Choosing colors |
| 96 | 99 | ~~~~~~~~~~~~~~~~ |
| 97 | 100 | |
| 98 | One can also modify the colors used by ``plot`` with the ``cmap`` option (for a full list of colormaps, see the `matplotlib website <http://matplotlib.org/users/colormaps.html>`_): | |
| 101 | One can also modify the colors used by :meth:`~GeoDataFrame.plot` with the ``cmap`` option (for a full list of colormaps, see the `matplotlib website <http://matplotlib.org/users/colormaps.html>`_): | |
| 99 | 102 | |
| 100 | 103 | .. ipython:: python |
| 101 | 104 | |
| 153 | 156 | }, |
| 154 | 157 | ); |
| 155 | 158 | |
| 159 | Other map customizations | |
| 160 | ~~~~~~~~~~~~~~~~~~~~~~~~ | |
| 161 | ||
| 162 | Maps usually do not have to have axis labels. You can turn them off using ``set_axis_off()`` or ``axis("off")`` axis methods. | |
| 163 | ||
| 164 | .. ipython:: python | |
| 165 | ||
| 166 | ax = world.plot() | |
| 167 | @savefig set_axis_off.png | |
| 168 | ax.set_axis_off(); | |
| 169 | ||
| 156 | 170 | Maps with Layers |
| 157 | 171 | ----------------- |
| 158 | 172 | |
| 10 | 10 | |
| 11 | 11 | There are two ways to combine datasets in *geopandas* -- attribute joins and spatial joins. |
| 12 | 12 | |
| 13 | In an attribute join, a ``GeoSeries`` or ``GeoDataFrame`` is combined with a regular *pandas* ``Series`` or ``DataFrame`` based on a common variable. This is analogous to normal merging or joining in *pandas*. | |
| 13 | In an attribute join, a :class:`GeoSeries` or :class:`GeoDataFrame` is | |
| 14 | combined with a regular :class:`pandas.Series` or :class:`pandas.DataFrame` based on a | |
| 15 | common variable. This is analogous to normal merging or joining in *pandas*. | |
| 14 | 16 | |
| 15 | In a Spatial Join, observations from two ``GeoSeries`` or ``GeoDataFrames`` are combined based on their spatial relationship to one another. | |
| 17 | In a Spatial Join, observations from two :class:`GeoSeries` or :class:`GeoDataFrame` | |
| 18 | are combined based on their spatial relationship to one another. | |
| 16 | 19 | |
| 17 | 20 | In the following examples, we use these datasets: |
| 18 | 21 | |
| 33 | 36 | Appending |
| 34 | 37 | --------- |
| 35 | 38 | |
| 36 | Appending GeoDataFrames and GeoSeries uses pandas ``append`` methods. Keep in mind, that appended geometry columns needs to have the same CRS. | |
| 39 | Appending :class:`GeoDataFrame` and :class:`GeoSeries` uses pandas :meth:`~pandas.DataFrame.append` methods. | |
| 40 | Keep in mind, that appended geometry columns needs to have the same CRS. | |
| 37 | 41 | |
| 38 | 42 | .. ipython:: python |
| 39 | 43 | |
| 49 | 53 | Attribute Joins |
| 50 | 54 | ---------------- |
| 51 | 55 | |
| 52 | Attribute joins are accomplished using the ``merge`` method. In general, it is recommended to use the ``merge`` method called from the spatial dataset. With that said, the stand-alone ``merge`` function will work if the GeoDataFrame is in the ``left`` argument; if a DataFrame is in the ``left`` argument and a GeoDataFrame is in the ``right`` position, the result will no longer be a GeoDataFrame. | |
| 56 | Attribute joins are accomplished using the :meth:`~pandas.DataFrame.merge` method. In general, it is recommended | |
| 57 | to use the ``merge()`` method called from the spatial dataset. With that said, the stand-alone | |
| 58 | :func:`pandas.merge` function will work if the :class:`GeoDataFrame` is in the ``left`` argument; | |
| 59 | if a :class:`~pandas.DataFrame` is in the ``left`` argument and a :class:`GeoDataFrame` | |
| 60 | is in the ``right`` position, the result will no longer be a :class:`GeoDataFrame`. | |
| 53 | 61 | |
| 54 | ||
| 55 | For example, consider the following merge that adds full names to a ``GeoDataFrame`` that initially has only ISO codes for each country by merging it with a *pandas* ``DataFrame``. | |
| 62 | For example, consider the following merge that adds full names to a :class:`GeoDataFrame` | |
| 63 | that initially has only ISO codes for each country by merging it with a :class:`~pandas.DataFrame`. | |
| 56 | 64 | |
| 57 | 65 | .. ipython:: python |
| 58 | 66 | |
| 65 | 73 | # Merge with `merge` method on shared variable (iso codes): |
| 66 | 74 | country_shapes = country_shapes.merge(country_names, on='iso_a3') |
| 67 | 75 | country_shapes.head() |
| 68 | ||
| 69 | 76 | |
| 70 | 77 | |
| 71 | 78 | Spatial Joins |
| 83 | 90 | |
| 84 | 91 | # Execute spatial join |
| 85 | 92 | |
| 86 | cities_with_country = geopandas.sjoin(cities, countries, how="inner", op='intersects') | |
| 93 | cities_with_country = cities.sjoin(countries, how="inner", predicate='intersects') | |
| 87 | 94 | cities_with_country.head() |
| 88 | 95 | |
| 89 | 96 | |
| 90 | Sjoin Arguments | |
| 91 | ~~~~~~~~~~~~~~~~ | |
| 97 | GeoPandas provides two spatial-join functions: | |
| 92 | 98 | |
| 93 | ``sjoin()`` has two core arguments: ``how`` and ``op``. | |
| 99 | - :meth:`GeoDataFrame.sjoin`: joins based on binary predicates (intersects, contains, etc.) | |
| 100 | - :meth:`GeoDataFrame.sjoin_nearest`: joins based on proximity, with the ability to set a maximum search radius. | |
| 94 | 101 | |
| 95 | **op** | |
| 102 | .. note:: | |
| 103 | For historical reasons, both methods are also available as top-level functions :func:`sjoin` and :func:`sjoin_nearest`. | |
| 104 | It is recommended to use methods as the functions may be deprecated in the future. | |
| 96 | 105 | |
| 97 | The ``op`` argument specifies how ``geopandas`` decides whether or not to join the attributes of one object to another, based on their geometric relationship. | |
| 106 | Binary Predicate Joins | |
| 107 | ~~~~~~~~~~~~~~~~~~~~~~ | |
| 98 | 108 | |
| 99 | The values for ``op`` correspond to the names of geometric binary predicates and depend on the spatial index implementation. | |
| 109 | Binary predicate joins are available via :meth:`GeoDataFrame.sjoin`. | |
| 100 | 110 | |
| 101 | The default spatial index in GeoPandas currently supports the following values for ``op``: | |
| 111 | :meth:`GeoDataFrame.sjoin` has two core arguments: ``how`` and ``predicate``. | |
| 112 | ||
| 113 | **predicate** | |
| 114 | ||
| 115 | The ``predicate`` argument specifies how ``geopandas`` decides whether or not to join the attributes of one | |
| 116 | object to another, based on their geometric relationship. | |
| 117 | ||
| 118 | The values for ``predicate`` correspond to the names of geometric binary predicates and depend on the spatial | |
| 119 | index implementation. | |
| 120 | ||
| 121 | The default spatial index in ``geopandas`` currently supports the following values for ``predicate`` which are | |
| 122 | defined in the | |
| 123 | `Shapely documentation <http://shapely.readthedocs.io/en/latest/manual.html#binary-predicates>`__: | |
| 102 | 124 | |
| 103 | 125 | * `intersects` |
| 104 | 126 | * `contains` |
| 107 | 129 | * `crosses` |
| 108 | 130 | * `overlaps` |
| 109 | 131 | |
| 110 | You can read more about each join type in the `Shapely documentation <http://shapely.readthedocs.io/en/latest/manual.html#binary-predicates>`__. | |
| 111 | ||
| 112 | 132 | **how** |
| 113 | 133 | |
| 114 | The `how` argument specifies the type of join that will occur and which geometry is retained in the resultant geodataframe. It accepts the following options: | |
| 134 | The `how` argument specifies the type of join that will occur and which geometry is retained in the resultant | |
| 135 | :class:`GeoDataFrame`. It accepts the following options: | |
| 115 | 136 | |
| 116 | * ``left``: use the index from the first (or `left_df`) geodataframe that you provide to ``sjoin``; retain only the `left_df` geometry column | |
| 137 | * ``left``: use the index from the first (or `left_df`) :class:`GeoDataFrame` that you provide | |
| 138 | to :meth:`GeoDataFrame.sjoin`; retain only the `left_df` geometry column | |
| 117 | 139 | * ``right``: use index from second (or `right_df`); retain only the `right_df` geometry column |
| 118 | * ``inner``: use intersection of index values from both geodataframes; retain only the `left_df` geometry column | |
| 140 | * ``inner``: use intersection of index values from both :class:`GeoDataFrame`; retain only the `left_df` geometry column | |
| 119 | 141 | |
| 120 | Note more complicated spatial relationships can be studied by combining geometric operations with spatial join. To find all polygons within a given distance of a point, for example, one can first use the ``buffer`` method to expand each point into a circle of appropriate radius, then intersect those buffered circles with the polygons in question. | |
| 142 | Note more complicated spatial relationships can be studied by combining geometric operations with spatial join. | |
| 143 | To find all polygons within a given distance of a point, for example, one can first use the :meth:`~geopandas.GeoSeries.buffer` method to expand each | |
| 144 | point into a circle of appropriate radius, then intersect those buffered circles with the polygons in question. | |
| 145 | ||
| 146 | Nearest Joins | |
| 147 | ~~~~~~~~~~~~~ | |
| 148 | ||
| 149 | Proximity-based joins can be done via :meth:`GeoDataFrame.sjoin_nearest`. | |
| 150 | ||
| 151 | :meth:`GeoDataFrame.sjoin_nearest` shares the ``how`` argument with :meth:`GeoDataFrame.sjoin`, and | |
| 152 | includes two additional arguments: ``max_distance`` and ``distance_col``. | |
| 153 | ||
| 154 | **max_distance** | |
| 155 | ||
| 156 | The ``max_distance`` argument specifies a maximum search radius for matching geometries. This can have a considerable performance impact in some cases. | |
| 157 | If you can, it is highly recommended that you use this parameter. | |
| 158 | ||
| 159 | **distance_col** | |
| 160 | ||
| 161 | If set, the resultant GeoDataFrame will include a column with this name containing the computed distances between an input geometry and the nearest geometry. | |
| 21 | 21 | a Shapely geometry object. |
| 22 | 22 | - **Missing geometries** are unknown values in a GeoSeries. They will typically |
| 23 | 23 | be propagated in operations (for example in calculations of the area or of |
| 24 | the intersection), or ignored in reductions such as ``unary_union``. | |
| 24 | the intersection), or ignored in reductions such as :attr:`~GeoSeries.unary_union`. | |
| 25 | 25 | The scalar object (when accessing a single element of a GeoSeries) is the |
| 26 | 26 | Python ``None`` object. |
| 27 | 27 | |
| 66 | 66 | |
| 67 | 67 | s.is_empty |
| 68 | 68 | |
| 69 | To get only the actual geometry objects that are neiter missing nor empty, | |
| 69 | To get only the actual geometry objects that are neither missing nor empty, | |
| 70 | 70 | you can use a combination of both: |
| 71 | 71 | |
| 72 | 72 | .. ipython:: python |
| 159 | 159 | |
| 160 | 160 | .. code-block:: python |
| 161 | 161 | |
| 162 | >>> s1.intersection(s2) | |
| 162 | >>> s1.intersection(s2) | |
| 163 | 163 | 0 GEOMETRYCOLLECTION EMPTY |
| 164 | 164 | 1 POINT (1 1) |
| 165 | 165 | 2 GEOMETRYCOLLECTION EMPTY |
| 168 | 168 | * Starting from GeoPandas v0.6.0, :meth:`GeoSeries.align` will use missing |
| 169 | 169 | values to fill in the non-aligned indices, to be consistent with the |
| 170 | 170 | behaviour in pandas: |
| 171 | ||
| 171 | ||
| 172 | 172 | .. ipython:: python |
| 173 | 173 | |
| 174 | 174 | s1_aligned, s2_aligned = s1.align(s2) |
| 180 | 180 | depending on the spatial operation: |
| 181 | 181 | |
| 182 | 182 | .. ipython:: python |
| 183 | :okwarning: | |
| 183 | 184 | |
| 184 | 185 | s1.intersection(s2) |
| 29 | 29 | - CRS WKT string |
| 30 | 30 | - An authority string (i.e. "epsg:4326") |
| 31 | 31 | - An EPSG integer code (i.e. 4326) |
| 32 | - A ``pyproj.CRS`` | |
| 32 | - A :class:`pyproj.CRS <pyproj.crs.CRS>` | |
| 33 | 33 | - An object with a to_wkt method. |
| 34 | 34 | - PROJ string |
| 35 | 35 | - Dictionary of PROJ parameters |
| 85 | 85 | world = geopandas.read_file(geopandas.datasets.get_path('naturalearth_lowres')) |
| 86 | 86 | |
| 87 | 87 | # Check original projection |
| 88 | # (it's Platte Carre! x-y are long and lat) | |
| 88 | # (it's Plate Carrée! x-y are long and lat) | |
| 89 | 89 | world.crs |
| 90 | 90 | |
| 91 | 91 | # Visualize |
| 144 | 144 | --------------------------------------------------------- |
| 145 | 145 | |
| 146 | 146 | Starting with GeoPandas 0.7, the `.crs` attribute of a GeoSeries or GeoDataFrame |
| 147 | stores the CRS information as a ``pyproj.CRS``, and no longer as a proj4 string | |
| 147 | stores the CRS information as a :class:`pyproj.CRS <pyproj.crs.CRS>`, and no longer as a proj4 string | |
| 148 | 148 | or dict. |
| 149 | 149 | |
| 150 | 150 | Before, you might have seen this: |
| 175 | 175 | migration issues. |
| 176 | 176 | |
| 177 | 177 | See the `pyproj docs <https://pyproj4.github.io/pyproj/stable/>`__ for more on |
| 178 | the ``pyproj.CRS`` object. | |
| 178 | the :class:`pyproj.CRS <pyproj.crs.CRS>` object. | |
| 179 | 179 | |
| 180 | 180 | Importing data from files |
| 181 | 181 | ^^^^^^^^^^^^^^^^^^^^^^^^^ |
| 266 | 266 | **Other formats** |
| 267 | 267 | |
| 268 | 268 | Next to the EPSG code mentioned above, there are also other ways to specify the |
| 269 | CRS: an actual ``pyproj.CRS`` object, a WKT string, a PROJ JSON string, etc. | |
| 270 | Anything that is accepted by ``pyproj.CRS.from_user_input`` can by specified | |
| 269 | CRS: an actual :class:`pyproj.CRS <pyproj.crs.CRS>` object, a WKT string, a PROJ JSON string, etc. | |
| 270 | Anything that is accepted by :meth:`pyproj.CRS.from_user_input() <pyproj.crs.CRS.from_user_input>` can by specified | |
| 271 | 271 | to the ``crs`` keyword/attribute in GeoPandas. |
| 272 | 272 | |
| 273 | Also compatible CRS objects, such as from the ``rasterio`` package, can be | |
| 273 | Also compatible CRS objects, such as from the :mod:`rasterio` package, can be | |
| 274 | 274 | passed directly to GeoPandas. |
| 275 | 275 | |
| 276 | 276 | |
| 305 | 305 | There are many file sources and CRS definitions out there "in the wild" that |
| 306 | 306 | might have a CRS description that does not fully conform to the new standards of |
| 307 | 307 | PROJ > 6 (proj4 strings, older WKT formats, ...). In such cases, you will get a |
| 308 | ``pyproj.CRS`` object that might not be fully what you expected (e.g. not equal | |
| 308 | :class:`pyproj.CRS <pyproj.crs.CRS>` object that might not be fully what you expected (e.g. not equal | |
| 309 | 309 | to the expected EPSG code). Below we list a few possible cases. |
| 310 | 310 | |
| 311 | 311 | I get a "Bound CRS"? |
| 446 | 446 | ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^ |
| 447 | 447 | |
| 448 | 448 | If you relied on the ``.crs`` object being a dict or a string, such code can |
| 449 | be broken given it is now a ``pyproj.CRS`` object. But this object actually | |
| 449 | be broken given it is now a :class:`pyproj.CRS <pyproj.crs.CRS>` object. But this object actually | |
| 450 | 450 | provides a more robust interface to get information about the CRS. |
| 451 | 451 | |
| 452 | 452 | For example, if you used the following code to get the EPSG code: |
| 456 | 456 | gdf.crs['init'] |
| 457 | 457 | |
| 458 | 458 | This will no longer work. To get the EPSG code from a ``crs`` object, you can use |
| 459 | the ``to_epsg()`` method. | |
| 459 | the :meth:`~pyproj.crs.CRS.to_epsg` method. | |
| 460 | 460 | |
| 461 | 461 | Or to check if a CRS was a certain UTM zone: |
| 462 | 462 | |
| 470 | 470 | |
| 471 | 471 | gdf.crs.utm_zone is not None |
| 472 | 472 | |
| 473 | And there are many other methods available on the ``pyproj.CRS`` class to get | |
| 473 | And there are many other methods available on the :class:`pyproj.CRS <pyproj.crs.CRS>` class to get | |
| 474 | 474 | information about the CRS. |
| 0 | Re-projecting using GDAL with Rasterio and Fiona | |
| 1 | ================================================ | |
| 2 | ||
| 3 | The simplest method of re-projecting is :meth:`GeoDataFrame.to_crs`. | |
| 4 | It uses ``pyproj`` as the engine and transforms the points within the geometries. | |
| 5 | ||
| 6 | These examples demonstrate how to use ``Fiona`` or ``rasterio`` as the engine to re-project your data. | |
| 7 | Fiona and rasterio are powered by GDAL and with algorithms that consider the geometry instead of | |
| 8 | just the points the geometry contains. This is particularly useful for antimeridian cutting. | |
| 9 | However, this also means the transformation is not as fast. | |
| 10 | ||
| 11 | ||
| 12 | Fiona Example | |
| 13 | ------------- | |
| 14 | ||
| 15 | .. code-block:: python | |
| 16 | ||
| 17 | from functools import partial | |
| 18 | ||
| 19 | import fiona | |
| 20 | import geopandas | |
| 21 | from fiona.transform import transform_geom | |
| 22 | from packaging import version | |
| 23 | from pyproj import CRS | |
| 24 | from pyproj.enums import WktVersion | |
| 25 | from shapely.geometry import mapping, shape | |
| 26 | ||
| 27 | ||
| 28 | # set up Fiona transformer | |
| 29 | def crs_to_fiona(proj_crs): | |
| 30 | proj_crs = CRS.from_user_input(proj_crs) | |
| 31 | if version.parse(fiona.__gdal_version__) < version.parse("3.0.0"): | |
| 32 | fio_crs = proj_crs.to_wkt(WktVersion.WKT1_GDAL) | |
| 33 | else: | |
| 34 | # GDAL 3+ can use WKT2 | |
| 35 | fio_crs = proj_crs.to_wkt() | |
| 36 | return fio_crs | |
| 37 | ||
| 38 | def base_transformer(geom, src_crs, dst_crs): | |
| 39 | return shape( | |
| 40 | transform_geom( | |
| 41 | src_crs=crs_to_fiona(src_crs), | |
| 42 | dst_crs=crs_to_fiona(dst_crs), | |
| 43 | geom=mapping(geom), | |
| 44 | antimeridian_cutting=True, | |
| 45 | ) | |
| 46 | ) | |
| 47 | ||
| 48 | # load example data | |
| 49 | world = geopandas.read_file(geopandas.datasets.get_path('naturalearth_lowres')) | |
| 50 | ||
| 51 | destination_crs = "EPSG:3395" | |
| 52 | forward_transformer = partial(base_transformer, src_crs=world.crs, dst_crs=destination_crs) | |
| 53 | ||
| 54 | # Reproject to Mercator (after dropping Antartica) | |
| 55 | world = world[(world.name != "Antarctica") & (world.name != "Fr. S. Antarctic Lands")] | |
| 56 | with fiona.Env(OGR_ENABLE_PARTIAL_REPROJECTION="YES"): | |
| 57 | mercator_world = world.set_geometry(world.geometry.apply(forward_transformer), crs=destination_crs) | |
| 58 | ||
| 59 | ||
| 60 | Rasterio Example | |
| 61 | ---------------- | |
| 62 | ||
| 63 | This example requires rasterio 1.2+ and GDAL 3+. | |
| 64 | ||
| 65 | ||
| 66 | .. code-block:: python | |
| 67 | ||
| 68 | import geopandas | |
| 69 | import rasterio.warp | |
| 70 | from shapely.geometry import shape | |
| 71 | ||
| 72 | # load example data | |
| 73 | world = geopandas.read_file(geopandas.datasets.get_path('naturalearth_lowres')) | |
| 74 | # Reproject to Mercator (after dropping Antartica) | |
| 75 | world = world[(world.name != "Antarctica") & (world.name != "Fr. S. Antarctic Lands")] | |
| 76 | ||
| 77 | destination_crs = "EPSG:3395" | |
| 78 | geometry = rasterio.warp.transform_geom( | |
| 79 | src_crs=world.crs, | |
| 80 | dst_crs=destination_crs, | |
| 81 | geom=world.geometry.values, | |
| 82 | ) | |
| 83 | mercator_world = world.set_geometry( | |
| 84 | [shape(geom) for geom in geometry], | |
| 85 | crs=destination_crs, | |
| 86 | ) |
| 0 | .. currentmodule:: geopandas | |
| 1 | ||
| 0 | 2 | .. ipython:: python |
| 1 | 3 | :suppress: |
| 2 | 4 | |
| 13 | 15 | those datasets overlap (or don't overlap). These manipulations are often |
| 14 | 16 | referred using the language of sets -- intersections, unions, and differences. |
| 15 | 17 | These types of operations are made available in the *geopandas* library through |
| 16 | the ``overlay`` function. | |
| 18 | the :meth:`~geopandas.GeoDataFrame.overlay` method. | |
| 17 | 19 | |
| 18 | 20 | The basic idea is demonstrated by the graphic below but keep in mind that |
| 19 | 21 | overlays operate at the DataFrame level, not on individual geometries, and the |
| 20 | properties from both are retained. In effect, for every shape in the first | |
| 21 | GeoDataFrame, this operation is executed against every other shape in the other | |
| 22 | GeoDataFrame: | |
| 22 | properties from both are retained. In effect, for every shape in the left | |
| 23 | :class:`~geopandas.GeoDataFrame`, this operation is executed against every other shape in the right | |
| 24 | :class:`~geopandas.GeoDataFrame`: | |
| 23 | 25 | |
| 24 | 26 | .. image:: ../../_static/overlay_operations.png |
| 25 | 27 | |
| 26 | 28 | **Source: QGIS Documentation** |
| 27 | 29 | |
| 28 | (Note to users familiar with the *shapely* library: ``overlay`` can be thought | |
| 29 | of as offering versions of the standard *shapely* set-operations that deal with | |
| 30 | the complexities of applying set operations to two *GeoSeries*. The standard | |
| 31 | *shapely* set-operations are also available as ``GeoSeries`` methods.) | |
| 30 | .. note:: | |
| 31 | Note to users familiar with the *shapely* library: :meth:`~geopandas.GeoDataFrame.overlay` can be thought | |
| 32 | of as offering versions of the standard *shapely* set-operations that deal with | |
| 33 | the complexities of applying set operations to two *GeoSeries*. The standard | |
| 34 | *shapely* set-operations are also available as :class:`~geopandas.GeoSeries` methods. | |
| 32 | 35 | |
| 33 | 36 | |
| 34 | 37 | The different Overlay operations |
| 56 | 59 | df2.plot(ax=ax, color='green', alpha=0.5); |
| 57 | 60 | |
| 58 | 61 | We illustrate the different overlay modes with the above example. |
| 59 | The ``overlay`` function will determine the set of all individual geometries | |
| 62 | The :meth:`~geopandas.GeoDataFrame.overlay` method will determine the set of all individual geometries | |
| 60 | 63 | from overlaying the two input GeoDataFrames. This result covers the area covered |
| 61 | 64 | by the two input GeoDataFrames, and also preserves all unique regions defined by |
| 62 | 65 | the combined boundaries of the two GeoDataFrames. |
| 63 | 66 | |
| 67 | .. note:: | |
| 68 | For historical reasons, the overlay method is also available as a top-level function :func:`overlay`. | |
| 69 | It is recommended to use the method as the function may be deprecated in the future. | |
| 70 | ||
| 64 | 71 | When using ``how='union'``, all those possible geometries are returned: |
| 65 | 72 | |
| 66 | 73 | .. ipython:: python |
| 67 | 74 | |
| 68 | res_union = geopandas.overlay(df1, df2, how='union') | |
| 75 | res_union = df1.overlay(df2, how='union') | |
| 69 | 76 | res_union |
| 70 | 77 | |
| 71 | 78 | ax = res_union.plot(alpha=0.5, cmap='tab10') |
| 79 | 86 | |
| 80 | 87 | .. ipython:: python |
| 81 | 88 | |
| 82 | res_intersection = geopandas.overlay(df1, df2, how='intersection') | |
| 89 | res_intersection = df1.overlay(df2, how='intersection') | |
| 83 | 90 | res_intersection |
| 84 | 91 | |
| 85 | 92 | ax = res_intersection.plot(cmap='tab10') |
| 92 | 99 | |
| 93 | 100 | .. ipython:: python |
| 94 | 101 | |
| 95 | res_symdiff = geopandas.overlay(df1, df2, how='symmetric_difference') | |
| 102 | res_symdiff = df1.overlay(df2, how='symmetric_difference') | |
| 96 | 103 | res_symdiff |
| 97 | 104 | |
| 98 | 105 | ax = res_symdiff.plot(cmap='tab10') |
| 105 | 112 | |
| 106 | 113 | .. ipython:: python |
| 107 | 114 | |
| 108 | res_difference = geopandas.overlay(df1, df2, how='difference') | |
| 115 | res_difference = df1.overlay(df2, how='difference') | |
| 109 | 116 | res_difference |
| 110 | 117 | |
| 111 | 118 | ax = res_difference.plot(cmap='tab10') |
| 118 | 125 | |
| 119 | 126 | .. ipython:: python |
| 120 | 127 | |
| 121 | res_identity = geopandas.overlay(df1, df2, how='identity') | |
| 128 | res_identity = df1.overlay(df2, how='identity') | |
| 122 | 129 | res_identity |
| 123 | 130 | |
| 124 | 131 | ax = res_identity.plot(cmap='tab10') |
| 145 | 152 | countries = countries.to_crs('epsg:3395') |
| 146 | 153 | capitals = capitals.to_crs('epsg:3395') |
| 147 | 154 | |
| 148 | To illustrate the ``overlay`` function, consider the following case in which one | |
| 155 | To illustrate the :meth:`~geopandas.GeoDataFrame.overlay` method, consider the following case in which one | |
| 149 | 156 | wishes to identify the "core" portion of each country -- defined as areas within |
| 150 | 157 | 500km of a capital -- using a ``GeoDataFrame`` of countries and a |
| 151 | 158 | ``GeoDataFrame`` of capitals. |
| 170 | 177 | |
| 171 | 178 | .. ipython:: python |
| 172 | 179 | |
| 173 | country_cores = geopandas.overlay(countries, capitals, how='intersection') | |
| 180 | country_cores = countries.overlay(capitals, how='intersection') | |
| 174 | 181 | @savefig country_cores.png width=5in |
| 175 | 182 | country_cores.plot(alpha=0.5, edgecolor='k', cmap='tab10'); |
| 176 | 183 | |
| 178 | 185 | |
| 179 | 186 | .. ipython:: python |
| 180 | 187 | |
| 181 | country_peripheries = geopandas.overlay(countries, capitals, how='difference') | |
| 188 | country_peripheries = countries.overlay(capitals, how='difference') | |
| 182 | 189 | @savefig country_peripheries.png width=5in |
| 183 | 190 | country_peripheries.plot(alpha=0.5, edgecolor='k', cmap='tab10'); |
| 184 | 191 | |
| 193 | 200 | keep_geom_type keyword |
| 194 | 201 | ---------------------- |
| 195 | 202 | |
| 196 | In default settings, ``overlay`` returns only geometries of the same geometry type as df1 | |
| 203 | In default settings, :meth:`~geopandas.GeoDataFrame.overlay` returns only geometries of the same geometry type as GeoDataFrame | |
| 197 | 204 | (left one) has, where Polygon and MultiPolygon is considered as a same type (other types likewise). |
| 198 | 205 | You can control this behavior using ``keep_geom_type`` option, which is set to |
| 199 | 206 | True by default. Once set to False, ``overlay`` will return all geometry types resulting from |
| 204 | 211 | More Examples |
| 205 | 212 | ------------- |
| 206 | 213 | |
| 207 | A larger set of examples of the use of ``overlay`` can be found `here <http://nbviewer.jupyter.org/github/geopandas/geopandas/blob/master/examples/overlays.ipynb>`_ | |
| 214 | A larger set of examples of the use of :meth:`~geopandas.GeoDataFrame.overlay` can be found `here <https://nbviewer.jupyter.org/github/geopandas/geopandas/blob/master/doc/source/gallery/overlays.ipynb>`_ | |
| 208 | 215 | |
| 209 | 216 | |
| 210 | 217 | |
| 13 | 13 | Reading and Writing Files <user_guide/io> |
| 14 | 14 | Indexing and Selecting Data <user_guide/indexing> |
| 15 | 15 | Making Maps and plots <user_guide/mapping> |
| 16 | Interactive mapping <user_guide/interactive_mapping> | |
| 16 | 17 | Managing Projections <user_guide/projections> |
| 17 | 18 | Geometric Manipulations <user_guide/geometric_manipulations> |
| 18 | 19 | Set Operations with overlay <user_guide/set_operations> |
| 17 | 17 | "* Equal Intervals\n", |
| 18 | 18 | " - Separates the measure's interval into equal parts, 5C per bin.\n", |
| 19 | 19 | "* Natural Breaks (Fischer Jenks)\n", |
| 20 | " - This algorithm tries to split the rows into naturaly occurring clusters. The numbers per bin will depend on how the observations are located on the interval." | |
| 20 | " - This algorithm tries to split the rows into naturally occurring clusters. The numbers per bin will depend on how the observations are located on the interval." | |
| 21 | 21 | ] |
| 22 | 22 | }, |
| 23 | 23 | { |
| 8 | 8 | "\n", |
| 9 | 9 | "This example shows how to create a ``GeoDataFrame`` when starting from\n", |
| 10 | 10 | "a *regular* ``DataFrame`` that has coordinates either WKT\n", |
| 11 | "([well-known text](https://en.wikipedia.org/wiki/Well-known_text>))\n", | |
| 11 | "([well-known text](https://en.wikipedia.org/wiki/Well-known_text))\n", | |
| 12 | 12 | "format, or in\n", |
| 13 | 13 | "two columns.\n" |
| 14 | 14 | ] |
| 224 | 224 | }, |
| 225 | 225 | "nbformat": 4, |
| 226 | 226 | "nbformat_minor": 4 |
| 227 | }⏎ | |
| 227 | } | |
| 0 | { | |
| 1 | "cells": [ | |
| 2 | { | |
| 3 | "cell_type": "markdown", | |
| 4 | "metadata": {}, | |
| 5 | "source": [ | |
| 6 | "\n", | |
| 7 | "# Using GeoPandas with Rasterio to sample point data\n", | |
| 8 | "\n", | |
| 9 | "This example shows how to use GeoPandas with Rasterio. [Rasterio](https://rasterio.readthedocs.io/en/latest/index.html) is a package for reading and writing raster data.\n", | |
| 10 | "\n", | |
| 11 | "In this example a set of vector points is used to sample raster data at those points.\n", | |
| 12 | "\n", | |
| 13 | "The raster data used is Copernicus Sentinel data 2018 for Sentinel data.\n" | |
| 14 | ] | |
| 15 | }, | |
| 16 | { | |
| 17 | "cell_type": "code", | |
| 18 | "execution_count": null, | |
| 19 | "metadata": {}, | |
| 20 | "outputs": [], | |
| 21 | "source": [ | |
| 22 | "import geopandas\n", | |
| 23 | "import rasterio\n", | |
| 24 | "import matplotlib.pyplot as plt\n", | |
| 25 | "from shapely.geometry import Point" | |
| 26 | ] | |
| 27 | }, | |
| 28 | { | |
| 29 | "cell_type": "markdown", | |
| 30 | "metadata": {}, | |
| 31 | "source": [ | |
| 32 | "Create example vector data\n", | |
| 33 | "=============================\n", | |
| 34 | "\n", | |
| 35 | "Generate a geodataframe from a set of points\n" | |
| 36 | ] | |
| 37 | }, | |
| 38 | { | |
| 39 | "cell_type": "code", | |
| 40 | "execution_count": null, | |
| 41 | "metadata": {}, | |
| 42 | "outputs": [], | |
| 43 | "source": [ | |
| 44 | "# Create sampling points\n", | |
| 45 | "points = [Point(625466, 5621289), Point(626082, 5621627), Point(627116, 5621680), Point(625095, 5622358)]\n", | |
| 46 | "gdf = geopandas.GeoDataFrame([1, 2, 3, 4], geometry=points, crs=32630)" | |
| 47 | ] | |
| 48 | }, | |
| 49 | { | |
| 50 | "cell_type": "markdown", | |
| 51 | "metadata": {}, | |
| 52 | "source": [ | |
| 53 | "The ``GeoDataFrame`` looks like this:" | |
| 54 | ] | |
| 55 | }, | |
| 56 | { | |
| 57 | "cell_type": "code", | |
| 58 | "execution_count": null, | |
| 59 | "metadata": {}, | |
| 60 | "outputs": [], | |
| 61 | "source": [ | |
| 62 | "gdf.head()" | |
| 63 | ] | |
| 64 | }, | |
| 65 | { | |
| 66 | "cell_type": "markdown", | |
| 67 | "metadata": {}, | |
| 68 | "source": [ | |
| 69 | "Open the raster data\n", | |
| 70 | "=============================\n", | |
| 71 | "\n", | |
| 72 | "Use ``rasterio`` to open the raster data to be sampled" | |
| 73 | ] | |
| 74 | }, | |
| 75 | { | |
| 76 | "cell_type": "code", | |
| 77 | "execution_count": null, | |
| 78 | "metadata": {}, | |
| 79 | "outputs": [], | |
| 80 | "source": [ | |
| 81 | "src = rasterio.open('s2a_l2a_fishbourne.tif')" | |
| 82 | ] | |
| 83 | }, | |
| 84 | { | |
| 85 | "cell_type": "markdown", | |
| 86 | "metadata": {}, | |
| 87 | "source": [ | |
| 88 | "Let's see the raster data with the point data overlaid.\n", | |
| 89 | "\n" | |
| 90 | ] | |
| 91 | }, | |
| 92 | { | |
| 93 | "cell_type": "code", | |
| 94 | "execution_count": null, | |
| 95 | "metadata": { | |
| 96 | "tags": [ | |
| 97 | "nbsphinx-thumbnail" | |
| 98 | ] | |
| 99 | }, | |
| 100 | "outputs": [], | |
| 101 | "source": [ | |
| 102 | "from rasterio.plot import show\n", | |
| 103 | "\n", | |
| 104 | "fig, ax = plt.subplots()\n", | |
| 105 | "\n", | |
| 106 | "# transform rasterio plot to real world coords\n", | |
| 107 | "extent=[src.bounds[0], src.bounds[2], src.bounds[1], src.bounds[3]]\n", | |
| 108 | "ax = rasterio.plot.show(src, extent=extent, ax=ax, cmap='pink')\n", | |
| 109 | "\n", | |
| 110 | "gdf.plot(ax=ax)" | |
| 111 | ] | |
| 112 | }, | |
| 113 | { | |
| 114 | "cell_type": "markdown", | |
| 115 | "metadata": {}, | |
| 116 | "source": [ | |
| 117 | "Sampling the data\n", | |
| 118 | "===============\n", | |
| 119 | "Rasterio requires a list of the coordinates in x,y format rather than as the points that are in the geomentry column.\n", | |
| 120 | "\n", | |
| 121 | "This can be achieved using the code below" | |
| 122 | ] | |
| 123 | }, | |
| 124 | { | |
| 125 | "cell_type": "code", | |
| 126 | "execution_count": null, | |
| 127 | "metadata": {}, | |
| 128 | "outputs": [], | |
| 129 | "source": [ | |
| 130 | "coord_list = [(x,y) for x,y in zip(gdf['geometry'].x , gdf['geometry'].y)]" | |
| 131 | ] | |
| 132 | }, | |
| 133 | { | |
| 134 | "cell_type": "markdown", | |
| 135 | "metadata": {}, | |
| 136 | "source": [ | |
| 137 | "Carry out the sampling of the data and store the results in a new column called `value`. Note that if the image has more than one band, a value is returned for each band." | |
| 138 | ] | |
| 139 | }, | |
| 140 | { | |
| 141 | "cell_type": "code", | |
| 142 | "execution_count": null, | |
| 143 | "metadata": {}, | |
| 144 | "outputs": [], | |
| 145 | "source": [ | |
| 146 | "gdf['value'] = [x for x in src.sample(coord_list)]\n", | |
| 147 | "gdf.head()" | |
| 148 | ] | |
| 149 | } | |
| 150 | ], | |
| 151 | "metadata": { | |
| 152 | "kernelspec": { | |
| 153 | "display_name": "Python 3 (ipykernel)", | |
| 154 | "language": "python", | |
| 155 | "name": "python3" | |
| 156 | }, | |
| 157 | "language_info": { | |
| 158 | "codemirror_mode": { | |
| 159 | "name": "ipython", | |
| 160 | "version": 3 | |
| 161 | }, | |
| 162 | "file_extension": ".py", | |
| 163 | "mimetype": "text/x-python", | |
| 164 | "name": "python", | |
| 165 | "nbconvert_exporter": "python", | |
| 166 | "pygments_lexer": "ipython3", | |
| 167 | "version": "3.9.2" | |
| 168 | } | |
| 169 | }, | |
| 170 | "nbformat": 4, | |
| 171 | "nbformat_minor": 4 | |
| 172 | } |
| 0 | { | |
| 1 | "cells": [ | |
| 2 | { | |
| 3 | "cell_type": "markdown", | |
| 4 | "metadata": {}, | |
| 5 | "source": [ | |
| 6 | "# Adding a scale bar to a matplotlib plot\n", | |
| 7 | "When making a geospatial plot in matplotlib, you can use [maplotlib-scalebar library](https://pypi.org/project/matplotlib-scalebar/) to add a scale bar." | |
| 8 | ] | |
| 9 | }, | |
| 10 | { | |
| 11 | "cell_type": "code", | |
| 12 | "execution_count": null, | |
| 13 | "metadata": {}, | |
| 14 | "outputs": [], | |
| 15 | "source": [ | |
| 16 | "import geopandas as gpd\n", | |
| 17 | "from matplotlib_scalebar.scalebar import ScaleBar" | |
| 18 | ] | |
| 19 | }, | |
| 20 | { | |
| 21 | "cell_type": "markdown", | |
| 22 | "metadata": {}, | |
| 23 | "source": [ | |
| 24 | "## Creating a ScaleBar object\n", | |
| 25 | "The only required parameter for creating a ScaleBar object is `dx`. This is equal to a size of one pixel in real world. Value of this parameter depends on units of your CRS.\n", | |
| 26 | "\n", | |
| 27 | "### Projected coordinate system (meters)\n", | |
| 28 | "The easiest way to add a scale bar is using a projected coordinate system with meters as units. Just set `dx = 1`:" | |
| 29 | ] | |
| 30 | }, | |
| 31 | { | |
| 32 | "cell_type": "code", | |
| 33 | "execution_count": null, | |
| 34 | "metadata": { | |
| 35 | "tags": [ | |
| 36 | "nbsphinx-thumbnail" | |
| 37 | ] | |
| 38 | }, | |
| 39 | "outputs": [], | |
| 40 | "source": [ | |
| 41 | "nybb = gpd.read_file(gpd.datasets.get_path('nybb'))\n", | |
| 42 | "nybb = nybb.to_crs(32619) # Convert the dataset to a coordinate\n", | |
| 43 | "# system which uses meters\n", | |
| 44 | "\n", | |
| 45 | "ax = nybb.plot()\n", | |
| 46 | "ax.add_artist(ScaleBar(1))" | |
| 47 | ] | |
| 48 | }, | |
| 49 | { | |
| 50 | "cell_type": "markdown", | |
| 51 | "metadata": {}, | |
| 52 | "source": [ | |
| 53 | "### Geographic coordinate system (degrees)\n", | |
| 54 | "With a geographic coordinate system with degrees as units, `dx` should be equal to a distance in meters of two points with the same latitude (Y coordinate) which are one full degree of longitude (X) apart. You can calculate this distance by online calculator [(e.g. the Great Circle calculator)](http://edwilliams.org/gccalc.htm) or in geopandas.\\\n", | |
| 55 | "\\\n", | |
| 56 | "Firstly, we will create a GeoSeries with two points that have roughly the coordinates of NYC. They are located on the same latitude but one degree of longitude from each other. Their initial coordinates are specified in a geographic coordinate system (geographic WGS 84). They are then converted to a projected system for the calculation:" | |
| 57 | ] | |
| 58 | }, | |
| 59 | { | |
| 60 | "cell_type": "code", | |
| 61 | "execution_count": null, | |
| 62 | "metadata": {}, | |
| 63 | "outputs": [], | |
| 64 | "source": [ | |
| 65 | "from shapely.geometry.point import Point\n", | |
| 66 | "\n", | |
| 67 | "points = gpd.GeoSeries([Point(-73.5, 40.5), Point(-74.5, 40.5)], crs=4326) # Geographic WGS 84 - degrees\n", | |
| 68 | "points = points.to_crs(32619) # Projected WGS 84 - meters" | |
| 69 | ] | |
| 70 | }, | |
| 71 | { | |
| 72 | "cell_type": "markdown", | |
| 73 | "metadata": {}, | |
| 74 | "source": [ | |
| 75 | "After the conversion, we can calculate the distance between the points. The result slightly differs from the Great Circle Calculator but the difference is insignificant (84,921 and 84,767 meters):" | |
| 76 | ] | |
| 77 | }, | |
| 78 | { | |
| 79 | "cell_type": "code", | |
| 80 | "execution_count": null, | |
| 81 | "metadata": {}, | |
| 82 | "outputs": [], | |
| 83 | "source": [ | |
| 84 | "distance_meters = points[0].distance(points[1])" | |
| 85 | ] | |
| 86 | }, | |
| 87 | { | |
| 88 | "cell_type": "markdown", | |
| 89 | "metadata": {}, | |
| 90 | "source": [ | |
| 91 | "Finally, we are able to use geographic coordinate system in our plot. We set value of `dx` parameter to a distance we just calculated:" | |
| 92 | ] | |
| 93 | }, | |
| 94 | { | |
| 95 | "cell_type": "code", | |
| 96 | "execution_count": null, | |
| 97 | "metadata": { | |
| 98 | "scrolled": true | |
| 99 | }, | |
| 100 | "outputs": [], | |
| 101 | "source": [ | |
| 102 | "nybb = gpd.read_file(gpd.datasets.get_path('nybb'))\n", | |
| 103 | "nybb = nybb.to_crs(4326) # Using geographic WGS 84\n", | |
| 104 | "\n", | |
| 105 | "ax = nybb.plot()\n", | |
| 106 | "ax.add_artist(ScaleBar(distance_meters))" | |
| 107 | ] | |
| 108 | }, | |
| 109 | { | |
| 110 | "cell_type": "markdown", | |
| 111 | "metadata": {}, | |
| 112 | "source": [ | |
| 113 | "## Using other units \n", | |
| 114 | "The default unit for `dx` is m (meter). You can change this unit by the `units` and `dimension` parameters. There is a list of some possible `units` for various values of `dimension` below:\n", | |
| 115 | "\n", | |
| 116 | "| dimension | units |\n", | |
| 117 | "| ----- |:-----:|\n", | |
| 118 | "| si-length | km, m, cm, um|\n", | |
| 119 | "| imperial-length |in, ft, yd, mi|\n", | |
| 120 | "|si-length-reciprocal|1/m, 1/cm|\n", | |
| 121 | "|angle|deg|\n", | |
| 122 | "\n", | |
| 123 | "In the following example, we will leave the dataset in its initial CRS which uses feet as units. The plot shows scale of 2 leagues (approximately 11 kilometers):" | |
| 124 | ] | |
| 125 | }, | |
| 126 | { | |
| 127 | "cell_type": "code", | |
| 128 | "execution_count": null, | |
| 129 | "metadata": {}, | |
| 130 | "outputs": [], | |
| 131 | "source": [ | |
| 132 | "nybb = gpd.read_file(gpd.datasets.get_path('nybb'))\n", | |
| 133 | "\n", | |
| 134 | "ax = nybb.plot()\n", | |
| 135 | "ax.add_artist(ScaleBar(1, dimension=\"imperial-length\", units=\"ft\"))" | |
| 136 | ] | |
| 137 | }, | |
| 138 | { | |
| 139 | "cell_type": "markdown", | |
| 140 | "metadata": {}, | |
| 141 | "source": [ | |
| 142 | "## Customization of the scale bar" | |
| 143 | ] | |
| 144 | }, | |
| 145 | { | |
| 146 | "cell_type": "code", | |
| 147 | "execution_count": null, | |
| 148 | "metadata": { | |
| 149 | "scrolled": true | |
| 150 | }, | |
| 151 | "outputs": [], | |
| 152 | "source": [ | |
| 153 | "nybb = gpd.read_file(gpd.datasets.get_path('nybb')).to_crs(32619)\n", | |
| 154 | "ax = nybb.plot()\n", | |
| 155 | "\n", | |
| 156 | "# Position and layout\n", | |
| 157 | "scale1 = ScaleBar(\n", | |
| 158 | "dx=1, label='Scale 1',\n", | |
| 159 | " location='upper left', # in relation to the whole plot\n", | |
| 160 | " label_loc='left', scale_loc='bottom' # in relation to the line\n", | |
| 161 | ")\n", | |
| 162 | "\n", | |
| 163 | "# Color\n", | |
| 164 | "scale2 = ScaleBar(\n", | |
| 165 | " dx=1, label='Scale 2', location='center', \n", | |
| 166 | " color='#b32400', box_color='yellow',\n", | |
| 167 | " box_alpha=0.8 # Slightly transparent box\n", | |
| 168 | ")\n", | |
| 169 | "\n", | |
| 170 | "# Font and text formatting\n", | |
| 171 | "scale3 = ScaleBar(\n", | |
| 172 | " dx=1, label='Scale 3',\n", | |
| 173 | " font_properties={'family':'serif', 'size': 'large'}, # For more information, see the cell below\n", | |
| 174 | " scale_formatter=lambda value, unit: f'> {value} {unit} <'\n", | |
| 175 | ")\n", | |
| 176 | "\n", | |
| 177 | "ax.add_artist(scale1)\n", | |
| 178 | "ax.add_artist(scale2)\n", | |
| 179 | "ax.add_artist(scale3)" | |
| 180 | ] | |
| 181 | }, | |
| 182 | { | |
| 183 | "cell_type": "markdown", | |
| 184 | "metadata": {}, | |
| 185 | "source": [ | |
| 186 | "*Note:* Font is specified by six properties: `family`, `style`, `variant`, `stretch`, `weight`, `size` (and `math_fontfamily`). See [more](https://matplotlib.org/stable/api/font_manager_api.html#matplotlib.font_manager.FontProperties).\\\n", | |
| 187 | "\\\n", | |
| 188 | "For more information about matplotlib-scalebar library, see the [PyPI](https://pypi.org/project/matplotlib-scalebar/) or [GitHub](https://github.com/ppinard/matplotlib-scalebar) page." | |
| 189 | ] | |
| 190 | } | |
| 191 | ], | |
| 192 | "metadata": { | |
| 193 | "interpreter": { | |
| 194 | "hash": "9914e2881520d4f08a067c2c2c181121476026b863eca2e121cd0758701ab602" | |
| 195 | }, | |
| 196 | "kernelspec": { | |
| 197 | "display_name": "Python 3", | |
| 198 | "language": "python", | |
| 199 | "name": "python3" | |
| 200 | }, | |
| 201 | "language_info": { | |
| 202 | "codemirror_mode": { | |
| 203 | "name": "ipython", | |
| 204 | "version": 3 | |
| 205 | }, | |
| 206 | "file_extension": ".py", | |
| 207 | "mimetype": "text/x-python", | |
| 208 | "name": "python", | |
| 209 | "nbconvert_exporter": "python", | |
| 210 | "pygments_lexer": "ipython3", | |
| 211 | "version": "3.9.2" | |
| 212 | } | |
| 213 | }, | |
| 214 | "nbformat": 4, | |
| 215 | "nbformat_minor": 4 | |
| 216 | } |
| 1 | 1 | "cells": [ |
| 2 | 2 | { |
| 3 | 3 | "cell_type": "markdown", |
| 4 | "metadata": {}, | |
| 5 | 4 | "source": [ |
| 6 | 5 | "# Overlays\n", |
| 7 | 6 | "\n", |
| 15 | 14 | "not on individual geometries, and the properties from both are retained\n", |
| 16 | 15 | "\n", |
| 17 | 16 | "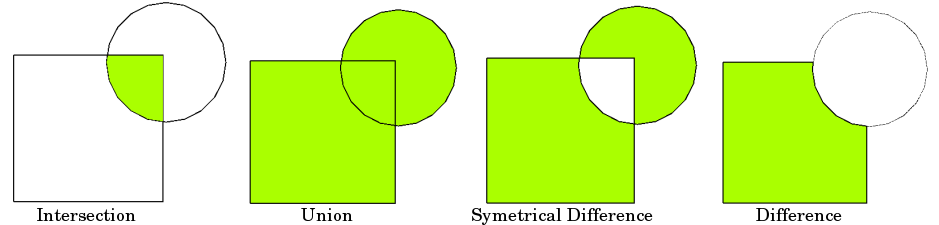" |
| 18 | ] | |
| 19 | }, | |
| 20 | { | |
| 21 | "cell_type": "markdown", | |
| 22 | "metadata": {}, | |
| 17 | ], | |
| 18 | "metadata": {} | |
| 19 | }, | |
| 20 | { | |
| 21 | "cell_type": "markdown", | |
| 23 | 22 | "source": [ |
| 24 | 23 | "Now we can load up two GeoDataFrames containing (multi)polygon geometries..." |
| 25 | ] | |
| 26 | }, | |
| 27 | { | |
| 28 | "cell_type": "code", | |
| 29 | "execution_count": null, | |
| 30 | "metadata": {}, | |
| 31 | "outputs": [], | |
| 24 | ], | |
| 25 | "metadata": {} | |
| 26 | }, | |
| 27 | { | |
| 28 | "cell_type": "code", | |
| 29 | "execution_count": null, | |
| 32 | 30 | "source": [ |
| 33 | 31 | "%matplotlib inline\n", |
| 34 | 32 | "from shapely.geometry import Point\n", |
| 46 | 44 | " {'geometry': Point(x, y).buffer(10000), 'value1': x + y, 'value2': x - y}\n", |
| 47 | 45 | " for x, y in zip(range(b[0], b[2], int((b[2] - b[0]) / N)),\n", |
| 48 | 46 | " range(b[1], b[3], int((b[3] - b[1]) / N)))])" |
| 49 | ] | |
| 50 | }, | |
| 51 | { | |
| 52 | "cell_type": "markdown", | |
| 53 | "metadata": {}, | |
| 47 | ], | |
| 48 | "outputs": [], | |
| 49 | "metadata": {} | |
| 50 | }, | |
| 51 | { | |
| 52 | "cell_type": "markdown", | |
| 54 | 53 | "source": [ |
| 55 | 54 | "The first dataframe contains multipolygons of the NYC boros" |
| 56 | ] | |
| 57 | }, | |
| 58 | { | |
| 59 | "cell_type": "code", | |
| 60 | "execution_count": null, | |
| 61 | "metadata": {}, | |
| 62 | "outputs": [], | |
| 55 | ], | |
| 56 | "metadata": {} | |
| 57 | }, | |
| 58 | { | |
| 59 | "cell_type": "code", | |
| 60 | "execution_count": null, | |
| 63 | 61 | "source": [ |
| 64 | 62 | "polydf.plot()" |
| 65 | ] | |
| 66 | }, | |
| 67 | { | |
| 68 | "cell_type": "markdown", | |
| 69 | "metadata": {}, | |
| 63 | ], | |
| 64 | "outputs": [], | |
| 65 | "metadata": {} | |
| 66 | }, | |
| 67 | { | |
| 68 | "cell_type": "markdown", | |
| 70 | 69 | "source": [ |
| 71 | 70 | "And the second GeoDataFrame is a sequentially generated set of circles in the same geographic space. We'll plot these with a [different color palette](https://matplotlib.org/examples/color/colormaps_reference.html)." |
| 72 | ] | |
| 73 | }, | |
| 74 | { | |
| 75 | "cell_type": "code", | |
| 76 | "execution_count": null, | |
| 77 | "metadata": {}, | |
| 78 | "outputs": [], | |
| 71 | ], | |
| 72 | "metadata": {} | |
| 73 | }, | |
| 74 | { | |
| 75 | "cell_type": "code", | |
| 76 | "execution_count": null, | |
| 79 | 77 | "source": [ |
| 80 | 78 | "polydf2.plot(cmap='tab20b')" |
| 81 | ] | |
| 82 | }, | |
| 83 | { | |
| 84 | "cell_type": "markdown", | |
| 85 | "metadata": {}, | |
| 79 | ], | |
| 80 | "outputs": [], | |
| 81 | "metadata": {} | |
| 82 | }, | |
| 83 | { | |
| 84 | "cell_type": "markdown", | |
| 86 | 85 | "source": [ |
| 87 | 86 | "The `geopandas.tools.overlay` function takes three arguments:\n", |
| 88 | 87 | "\n", |
| 98 | 97 | " 'symmetric_difference',\n", |
| 99 | 98 | " 'difference']\n", |
| 100 | 99 | "\n", |
| 101 | "So let's identify the areas (and attributes) where both dataframes intersect using the `overlay` tool. " | |
| 102 | ] | |
| 103 | }, | |
| 104 | { | |
| 105 | "cell_type": "code", | |
| 106 | "execution_count": null, | |
| 107 | "metadata": {}, | |
| 108 | "outputs": [], | |
| 109 | "source": [ | |
| 110 | "from geopandas.tools import overlay\n", | |
| 111 | "newdf = overlay(polydf, polydf2, how=\"intersection\")\n", | |
| 112 | "newdf.plot(cmap='tab20b')" | |
| 113 | ] | |
| 114 | }, | |
| 115 | { | |
| 116 | "cell_type": "markdown", | |
| 117 | "metadata": {}, | |
| 100 | "So let's identify the areas (and attributes) where both dataframes intersect using the `overlay` method. " | |
| 101 | ], | |
| 102 | "metadata": {} | |
| 103 | }, | |
| 104 | { | |
| 105 | "cell_type": "code", | |
| 106 | "execution_count": null, | |
| 107 | "source": [ | |
| 108 | "newdf = polydf.overlay(polydf2, how=\"intersection\")\n", | |
| 109 | "newdf.plot(cmap='tab20b')" | |
| 110 | ], | |
| 111 | "outputs": [], | |
| 112 | "metadata": {} | |
| 113 | }, | |
| 114 | { | |
| 115 | "cell_type": "markdown", | |
| 118 | 116 | "source": [ |
| 119 | 117 | "And take a look at the attributes; we see that the attributes from both of the original GeoDataFrames are retained. " |
| 120 | ] | |
| 121 | }, | |
| 122 | { | |
| 123 | "cell_type": "code", | |
| 124 | "execution_count": null, | |
| 125 | "metadata": {}, | |
| 126 | "outputs": [], | |
| 118 | ], | |
| 119 | "metadata": {} | |
| 120 | }, | |
| 121 | { | |
| 122 | "cell_type": "code", | |
| 123 | "execution_count": null, | |
| 127 | 124 | "source": [ |
| 128 | 125 | "polydf.head()" |
| 129 | ] | |
| 130 | }, | |
| 131 | { | |
| 132 | "cell_type": "code", | |
| 133 | "execution_count": null, | |
| 134 | "metadata": {}, | |
| 135 | "outputs": [], | |
| 126 | ], | |
| 127 | "outputs": [], | |
| 128 | "metadata": {} | |
| 129 | }, | |
| 130 | { | |
| 131 | "cell_type": "code", | |
| 132 | "execution_count": null, | |
| 136 | 133 | "source": [ |
| 137 | 134 | "polydf2.head()" |
| 138 | ] | |
| 139 | }, | |
| 140 | { | |
| 141 | "cell_type": "code", | |
| 142 | "execution_count": null, | |
| 143 | "metadata": {}, | |
| 144 | "outputs": [], | |
| 135 | ], | |
| 136 | "outputs": [], | |
| 137 | "metadata": {} | |
| 138 | }, | |
| 139 | { | |
| 140 | "cell_type": "code", | |
| 141 | "execution_count": null, | |
| 145 | 142 | "source": [ |
| 146 | 143 | "newdf.head()" |
| 147 | ] | |
| 148 | }, | |
| 149 | { | |
| 150 | "cell_type": "markdown", | |
| 151 | "metadata": {}, | |
| 144 | ], | |
| 145 | "outputs": [], | |
| 146 | "metadata": {} | |
| 147 | }, | |
| 148 | { | |
| 149 | "cell_type": "markdown", | |
| 152 | 150 | "source": [ |
| 153 | 151 | "Now let's look at the other `how` operations:" |
| 154 | ] | |
| 155 | }, | |
| 156 | { | |
| 157 | "cell_type": "code", | |
| 158 | "execution_count": null, | |
| 159 | "metadata": {}, | |
| 160 | "outputs": [], | |
| 161 | "source": [ | |
| 162 | "newdf = overlay(polydf, polydf2, how=\"union\")\n", | |
| 163 | "newdf.plot(cmap='tab20b')" | |
| 164 | ] | |
| 165 | }, | |
| 166 | { | |
| 167 | "cell_type": "code", | |
| 168 | "execution_count": null, | |
| 169 | "metadata": {}, | |
| 170 | "outputs": [], | |
| 171 | "source": [ | |
| 172 | "newdf = overlay(polydf, polydf2, how=\"identity\")\n", | |
| 173 | "newdf.plot(cmap='tab20b')" | |
| 174 | ] | |
| 175 | }, | |
| 176 | { | |
| 177 | "cell_type": "code", | |
| 178 | "execution_count": null, | |
| 152 | ], | |
| 153 | "metadata": {} | |
| 154 | }, | |
| 155 | { | |
| 156 | "cell_type": "code", | |
| 157 | "execution_count": null, | |
| 158 | "source": [ | |
| 159 | "newdf = polydf.overlay(polydf2, how=\"union\")\n", | |
| 160 | "newdf.plot(cmap='tab20b')" | |
| 161 | ], | |
| 162 | "outputs": [], | |
| 163 | "metadata": {} | |
| 164 | }, | |
| 165 | { | |
| 166 | "cell_type": "code", | |
| 167 | "execution_count": null, | |
| 168 | "source": [ | |
| 169 | "newdf = polydf.overlay(polydf2, how=\"identity\")\n", | |
| 170 | "newdf.plot(cmap='tab20b')" | |
| 171 | ], | |
| 172 | "outputs": [], | |
| 173 | "metadata": {} | |
| 174 | }, | |
| 175 | { | |
| 176 | "cell_type": "code", | |
| 177 | "execution_count": null, | |
| 178 | "source": [ | |
| 179 | "newdf = polydf.overlay(polydf2, how=\"symmetric_difference\")\n", | |
| 180 | "newdf.plot(cmap='tab20b')" | |
| 181 | ], | |
| 182 | "outputs": [], | |
| 179 | 183 | "metadata": { |
| 180 | 184 | "tags": [ |
| 181 | 185 | "nbsphinx-thumbnail" |
| 182 | 186 | ] |
| 183 | }, | |
| 184 | "outputs": [], | |
| 185 | "source": [ | |
| 186 | "newdf = overlay(polydf, polydf2, how=\"symmetric_difference\")\n", | |
| 187 | "newdf.plot(cmap='tab20b')" | |
| 188 | ] | |
| 189 | }, | |
| 190 | { | |
| 191 | "cell_type": "code", | |
| 192 | "execution_count": null, | |
| 193 | "metadata": {}, | |
| 194 | "outputs": [], | |
| 195 | "source": [ | |
| 196 | "newdf = overlay(polydf, polydf2, how=\"difference\")\n", | |
| 197 | "newdf.plot(cmap='tab20b')" | |
| 198 | ] | |
| 187 | } | |
| 188 | }, | |
| 189 | { | |
| 190 | "cell_type": "code", | |
| 191 | "execution_count": null, | |
| 192 | "source": [ | |
| 193 | "newdf = polydf.overlay(polydf2, how=\"difference\")\n", | |
| 194 | "newdf.plot(cmap='tab20b')" | |
| 195 | ], | |
| 196 | "outputs": [], | |
| 197 | "metadata": {} | |
| 199 | 198 | } |
| 200 | 199 | ], |
| 201 | 200 | "metadata": { |
| 202 | 201 | "kernelspec": { |
| 203 | "display_name": "Python 3", | |
| 202 | "display_name": "Python 3 (ipykernel)", | |
| 204 | 203 | "language": "python", |
| 205 | 204 | "name": "python3" |
| 206 | 205 | }, |
| 214 | 213 | "name": "python", |
| 215 | 214 | "nbconvert_exporter": "python", |
| 216 | 215 | "pygments_lexer": "ipython3", |
| 217 | "version": "3.9.1" | |
| 216 | "version": "3.9.2" | |
| 218 | 217 | } |
| 219 | 218 | }, |
| 220 | 219 | "nbformat": 4, |
| 221 | 220 | "nbformat_minor": 4 |
| 222 | } | |
| 221 | }⏎ | |
| 1 | 1 | "cells": [ |
| 2 | 2 | { |
| 3 | 3 | "cell_type": "markdown", |
| 4 | "metadata": {}, | |
| 5 | 4 | "source": [ |
| 6 | 5 | "# Clip Vector Data with GeoPandas\n", |
| 7 | 6 | "\n", |
| 8 | 7 | "\n", |
| 9 | 8 | "Learn how to clip geometries to the boundary of a polygon geometry\n", |
| 10 | 9 | "using GeoPandas." |
| 11 | ] | |
| 12 | }, | |
| 13 | { | |
| 14 | "cell_type": "markdown", | |
| 15 | "metadata": {}, | |
| 10 | ], | |
| 11 | "metadata": {} | |
| 12 | }, | |
| 13 | { | |
| 14 | "cell_type": "markdown", | |
| 16 | 15 | "source": [ |
| 17 | 16 | "The example below shows you how to clip a set of vector geometries\n", |
| 18 | 17 | "to the spatial extent / shape of another vector object. Both sets of geometries\n", |
| 32 | 31 | "be clipped to the total boundary of all polygons in clip object.\n", |
| 33 | 32 | "</div>\n", |
| 34 | 33 | "\n" |
| 35 | ] | |
| 36 | }, | |
| 37 | { | |
| 38 | "cell_type": "markdown", | |
| 39 | "metadata": {}, | |
| 34 | ], | |
| 35 | "metadata": {} | |
| 36 | }, | |
| 37 | { | |
| 38 | "cell_type": "markdown", | |
| 40 | 39 | "source": [ |
| 41 | 40 | "Import Packages\n", |
| 42 | 41 | "---------------\n", |
| 43 | 42 | "\n", |
| 44 | 43 | "To begin, import the needed packages.\n", |
| 45 | 44 | "\n" |
| 46 | ] | |
| 47 | }, | |
| 48 | { | |
| 49 | "cell_type": "code", | |
| 50 | "execution_count": null, | |
| 51 | "metadata": {}, | |
| 52 | "outputs": [], | |
| 45 | ], | |
| 46 | "metadata": {} | |
| 47 | }, | |
| 48 | { | |
| 49 | "cell_type": "code", | |
| 50 | "execution_count": null, | |
| 53 | 51 | "source": [ |
| 54 | 52 | "import matplotlib.pyplot as plt\n", |
| 55 | 53 | "import geopandas\n", |
| 56 | 54 | "from shapely.geometry import Polygon" |
| 57 | ] | |
| 58 | }, | |
| 59 | { | |
| 60 | "cell_type": "markdown", | |
| 61 | "metadata": {}, | |
| 55 | ], | |
| 56 | "outputs": [], | |
| 57 | "metadata": {} | |
| 58 | }, | |
| 59 | { | |
| 60 | "cell_type": "markdown", | |
| 62 | 61 | "source": [ |
| 63 | 62 | "Get or Create Example Data\n", |
| 64 | 63 | "--------------------------\n", |
| 67 | 66 | "Additionally, a polygon is created with shapely and then converted into a\n", |
| 68 | 67 | "GeoDataFrame with the same CRS as the GeoPandas world dataset.\n", |
| 69 | 68 | "\n" |
| 70 | ] | |
| 71 | }, | |
| 72 | { | |
| 73 | "cell_type": "code", | |
| 74 | "execution_count": null, | |
| 75 | "metadata": {}, | |
| 76 | "outputs": [], | |
| 69 | ], | |
| 70 | "metadata": {} | |
| 71 | }, | |
| 72 | { | |
| 73 | "cell_type": "code", | |
| 74 | "execution_count": null, | |
| 77 | 75 | "source": [ |
| 78 | 76 | "capitals = geopandas.read_file(geopandas.datasets.get_path(\"naturalearth_cities\"))\n", |
| 79 | 77 | "world = geopandas.read_file(geopandas.datasets.get_path(\"naturalearth_lowres\"))\n", |
| 84 | 82 | "# Create a custom polygon\n", |
| 85 | 83 | "polygon = Polygon([(0, 0), (0, 90), (180, 90), (180, 0), (0, 0)])\n", |
| 86 | 84 | "poly_gdf = geopandas.GeoDataFrame([1], geometry=[polygon], crs=world.crs)" |
| 87 | ] | |
| 88 | }, | |
| 89 | { | |
| 90 | "cell_type": "markdown", | |
| 91 | "metadata": {}, | |
| 85 | ], | |
| 86 | "outputs": [], | |
| 87 | "metadata": {} | |
| 88 | }, | |
| 89 | { | |
| 90 | "cell_type": "markdown", | |
| 92 | 91 | "source": [ |
| 93 | 92 | "Plot the Unclipped Data\n", |
| 94 | 93 | "-----------------------\n", |
| 95 | 94 | "\n" |
| 96 | ] | |
| 97 | }, | |
| 98 | { | |
| 99 | "cell_type": "code", | |
| 100 | "execution_count": null, | |
| 101 | "metadata": {}, | |
| 102 | "outputs": [], | |
| 95 | ], | |
| 96 | "metadata": {} | |
| 97 | }, | |
| 98 | { | |
| 99 | "cell_type": "code", | |
| 100 | "execution_count": null, | |
| 103 | 101 | "source": [ |
| 104 | 102 | "fig, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 8))\n", |
| 105 | 103 | "world.plot(ax=ax1)\n", |
| 111 | 109 | "ax1.set_axis_off()\n", |
| 112 | 110 | "ax2.set_axis_off()\n", |
| 113 | 111 | "plt.show()" |
| 114 | ] | |
| 115 | }, | |
| 116 | { | |
| 117 | "cell_type": "markdown", | |
| 118 | "metadata": {}, | |
| 112 | ], | |
| 113 | "outputs": [], | |
| 114 | "metadata": {} | |
| 115 | }, | |
| 116 | { | |
| 117 | "cell_type": "markdown", | |
| 119 | 118 | "source": [ |
| 120 | 119 | "Clip the Data\n", |
| 121 | 120 | "--------------\n", |
| 122 | 121 | "\n", |
| 123 | "When you call `clip`, the first object called is the object that will\n", | |
| 124 | "be clipped. The second object called is the clip extent. The returned output\n", | |
| 122 | "The object on which you call `clip` is the object that will\n", | |
| 123 | "be clipped. The object you pass is the clip extent. The returned output\n", | |
| 125 | 124 | "will be a new clipped GeoDataframe. All of the attributes for each returned\n", |
| 126 | 125 | "geometry will be retained when you clip.\n", |
| 127 | 126 | "\n", |
| 130 | 129 | "Note\n", |
| 131 | 130 | "\n", |
| 132 | 131 | "Recall that the data must be in the same CRS in order to use the\n", |
| 133 | "`clip` function. If the data are not in the same CRS, be sure to use\n", | |
| 132 | "`clip` method. If the data are not in the same CRS, be sure to use\n", | |
| 134 | 133 | "the GeoPandas `GeoDataFrame.to_crs` method to ensure both datasets\n", |
| 135 | 134 | "are in the same CRS.\n", |
| 136 | 135 | "</div>\n", |
| 137 | 136 | "\n" |
| 138 | ] | |
| 139 | }, | |
| 140 | { | |
| 141 | "cell_type": "markdown", | |
| 142 | "metadata": {}, | |
| 137 | ], | |
| 138 | "metadata": {} | |
| 139 | }, | |
| 140 | { | |
| 141 | "cell_type": "markdown", | |
| 143 | 142 | "source": [ |
| 144 | 143 | "Clip the World Data\n", |
| 145 | 144 | "--------------------\n", |
| 146 | 145 | "\n" |
| 147 | ] | |
| 148 | }, | |
| 149 | { | |
| 150 | "cell_type": "code", | |
| 151 | "execution_count": null, | |
| 152 | "metadata": { | |
| 153 | "tags": [ | |
| 154 | "nbsphinx-thumbnail" | |
| 155 | ] | |
| 156 | }, | |
| 157 | "outputs": [], | |
| 158 | "source": [ | |
| 159 | "world_clipped = geopandas.clip(world, polygon)\n", | |
| 146 | ], | |
| 147 | "metadata": {} | |
| 148 | }, | |
| 149 | { | |
| 150 | "cell_type": "code", | |
| 151 | "execution_count": null, | |
| 152 | "source": [ | |
| 153 | "world_clipped = world.clip(polygon)\n", | |
| 160 | 154 | "\n", |
| 161 | 155 | "# Plot the clipped data\n", |
| 162 | 156 | "# The plot below shows the results of the clip function applied to the world\n", |
| 168 | 162 | "ax.set_title(\"World Clipped\", fontsize=20)\n", |
| 169 | 163 | "ax.set_axis_off()\n", |
| 170 | 164 | "plt.show()" |
| 171 | ] | |
| 172 | }, | |
| 173 | { | |
| 174 | "cell_type": "markdown", | |
| 175 | "metadata": {}, | |
| 176 | "source": [ | |
| 165 | ], | |
| 166 | "outputs": [], | |
| 167 | "metadata": { | |
| 168 | "tags": [ | |
| 169 | "nbsphinx-thumbnail" | |
| 170 | ] | |
| 171 | } | |
| 172 | }, | |
| 173 | { | |
| 174 | "cell_type": "markdown", | |
| 175 | "source": [ | |
| 176 | "<div class=\"alert alert-info\">\n", | |
| 177 | " \n", | |
| 178 | "Note\n", | |
| 179 | "\n", | |
| 180 | "For historical reasons, the clip method is also available as a top-level function `geopandas.clip`.\n", | |
| 181 | "It is recommended to use the method as the function may be deprecated in the future.\n", | |
| 182 | "</div>\n", | |
| 183 | "\n", | |
| 177 | 184 | "Clip the Capitals Data\n", |
| 178 | 185 | "----------------------\n", |
| 179 | 186 | "\n" |
| 180 | ] | |
| 181 | }, | |
| 182 | { | |
| 183 | "cell_type": "code", | |
| 184 | "execution_count": null, | |
| 185 | "metadata": {}, | |
| 186 | "outputs": [], | |
| 187 | "source": [ | |
| 188 | "capitals_clipped = geopandas.clip(capitals, south_america)\n", | |
| 187 | ], | |
| 188 | "metadata": {} | |
| 189 | }, | |
| 190 | { | |
| 191 | "cell_type": "code", | |
| 192 | "execution_count": null, | |
| 193 | "source": [ | |
| 194 | "capitals_clipped = capitals.clip(south_america)\n", | |
| 189 | 195 | "\n", |
| 190 | 196 | "# Plot the clipped data\n", |
| 191 | 197 | "# The plot below shows the results of the clip function applied to the capital cities\n", |
| 195 | 201 | "ax.set_title(\"Capitals Clipped\", fontsize=20)\n", |
| 196 | 202 | "ax.set_axis_off()\n", |
| 197 | 203 | "plt.show()" |
| 198 | ] | |
| 204 | ], | |
| 205 | "outputs": [], | |
| 206 | "metadata": {} | |
| 199 | 207 | } |
| 200 | 208 | ], |
| 201 | 209 | "metadata": { |
| 219 | 227 | }, |
| 220 | 228 | "nbformat": 4, |
| 221 | 229 | "nbformat_minor": 4 |
| 222 | } | |
| 230 | }⏎ | |
| 9 | 9 | "This example shows how you can add a background basemap to plots created\n", |
| 10 | 10 | "with the geopandas ``.plot()`` method. This makes use of the\n", |
| 11 | 11 | "[contextily](https://github.com/geopandas/contextily) package to retrieve\n", |
| 12 | "web map tiles from several sources (OpenStreetMap, Stamen).\n" | |
| 13 | ] | |
| 14 | }, | |
| 15 | { | |
| 16 | "cell_type": "code", | |
| 17 | "execution_count": null, | |
| 18 | "metadata": {}, | |
| 19 | "outputs": [], | |
| 20 | "source": [ | |
| 21 | "import geopandas" | |
| 12 | "web map tiles from several sources (OpenStreetMap, Stamen). Also have a\n", | |
| 13 | "look at contextily's \n", | |
| 14 | "[introduction guide](https://contextily.readthedocs.io/en/latest/intro_guide.html#Using-transparent-layers)\n", | |
| 15 | "for possible new features not covered here.\n" | |
| 16 | ] | |
| 17 | }, | |
| 18 | { | |
| 19 | "cell_type": "code", | |
| 20 | "execution_count": null, | |
| 21 | "metadata": {}, | |
| 22 | "outputs": [], | |
| 23 | "source": [ | |
| 24 | "import geopandas\n", | |
| 25 | "import contextily as cx" | |
| 22 | 26 | ] |
| 23 | 27 | }, |
| 24 | 28 | { |
| 44 | 48 | "cell_type": "markdown", |
| 45 | 49 | "metadata": {}, |
| 46 | 50 | "source": [ |
| 47 | "Convert the data to Web Mercator\n", | |
| 48 | "================================\n", | |
| 49 | "\n", | |
| 51 | "## Matching coordinate systems \n", | |
| 52 | "\n", | |
| 53 | "\n", | |
| 54 | "Before adding web map tiles to this plot, we first need to ensure the\n", | |
| 55 | "coordinate reference systems (CRS) of the tiles and the data match.\n", | |
| 50 | 56 | "Web map tiles are typically provided in\n", |
| 51 | 57 | "[Web Mercator](https://en.wikipedia.org/wiki/Web_Mercator>)\n", |
| 52 | "([EPSG 3857](https://epsg.io/3857)), so we need to make sure to convert\n", | |
| 53 | "our data first to the same CRS to combine our polygons and background tiles\n", | |
| 54 | "in the same map:\n", | |
| 55 | "\n" | |
| 56 | ] | |
| 57 | }, | |
| 58 | { | |
| 59 | "cell_type": "code", | |
| 60 | "execution_count": null, | |
| 61 | "metadata": {}, | |
| 62 | "outputs": [], | |
| 63 | "source": [ | |
| 64 | "df = df.to_crs(epsg=3857)" | |
| 65 | ] | |
| 66 | }, | |
| 67 | { | |
| 68 | "cell_type": "code", | |
| 69 | "execution_count": null, | |
| 70 | "metadata": {}, | |
| 71 | "outputs": [], | |
| 72 | "source": [ | |
| 73 | "import contextily as ctx" | |
| 74 | ] | |
| 75 | }, | |
| 76 | { | |
| 77 | "cell_type": "markdown", | |
| 78 | "metadata": {}, | |
| 79 | "source": [ | |
| 80 | "Add background tiles to plot\n", | |
| 81 | "============================\n", | |
| 82 | "\n", | |
| 83 | "We can use `add_basemap` function of contextily to easily add a background\n", | |
| 84 | "map to our plot. :\n", | |
| 85 | "\n" | |
| 58 | "([EPSG 3857](https://epsg.io/3857)), so let us first check what\n", | |
| 59 | "CRS our NYC boroughs are in:" | |
| 60 | ] | |
| 61 | }, | |
| 62 | { | |
| 63 | "cell_type": "code", | |
| 64 | "execution_count": null, | |
| 65 | "metadata": {}, | |
| 66 | "outputs": [], | |
| 67 | "source": [ | |
| 68 | "df.crs" | |
| 69 | ] | |
| 70 | }, | |
| 71 | { | |
| 72 | "cell_type": "markdown", | |
| 73 | "metadata": {}, | |
| 74 | "source": [ | |
| 75 | "Now we know the CRS do not match, so we need to choose in which\n", | |
| 76 | "CRS we wish to visualize the data: either the CRS of the tiles,\n", | |
| 77 | "the one of the data, or even a different one.\n", | |
| 78 | "\n", | |
| 79 | "The first option to match CRS is to leverage the `to_crs` method\n", | |
| 80 | "of GeoDataFrames to convert the CRS of our data, here to Web Mercator:" | |
| 81 | ] | |
| 82 | }, | |
| 83 | { | |
| 84 | "cell_type": "code", | |
| 85 | "execution_count": null, | |
| 86 | "metadata": {}, | |
| 87 | "outputs": [], | |
| 88 | "source": [ | |
| 89 | "df_wm = df.to_crs(epsg=3857)" | |
| 90 | ] | |
| 91 | }, | |
| 92 | { | |
| 93 | "cell_type": "markdown", | |
| 94 | "metadata": {}, | |
| 95 | "source": [ | |
| 96 | "We can then use `add_basemap` function of contextily to easily add a\n", | |
| 97 | "background map to our plot:" | |
| 86 | 98 | ] |
| 87 | 99 | }, |
| 88 | 100 | { |
| 95 | 107 | }, |
| 96 | 108 | "outputs": [], |
| 97 | 109 | "source": [ |
| 110 | "ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 111 | "cx.add_basemap(ax)" | |
| 112 | ] | |
| 113 | }, | |
| 114 | { | |
| 115 | "cell_type": "markdown", | |
| 116 | "metadata": {}, | |
| 117 | "source": [ | |
| 118 | "If we want to convert the CRS of the tiles instead, which might be advisable\n", | |
| 119 | "for large datasets, we can use the `crs` keyword argument of `add_basemap`\n", | |
| 120 | "as follows:" | |
| 121 | ] | |
| 122 | }, | |
| 123 | { | |
| 124 | "cell_type": "code", | |
| 125 | "execution_count": null, | |
| 126 | "metadata": {}, | |
| 127 | "outputs": [], | |
| 128 | "source": [ | |
| 98 | 129 | "ax = df.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", |
| 99 | "ctx.add_basemap(ax)" | |
| 130 | "cx.add_basemap(ax, crs=df.crs)" | |
| 131 | ] | |
| 132 | }, | |
| 133 | { | |
| 134 | "cell_type": "markdown", | |
| 135 | "metadata": {}, | |
| 136 | "source": [ | |
| 137 | "This reprojects map tiles to a target CRS which may in some cases cause a\n", | |
| 138 | "loss of sharpness. See \n", | |
| 139 | "[contextily's guide on warping tiles](https://contextily.readthedocs.io/en/latest/warping_guide.html)\n", | |
| 140 | "for more information on the subject." | |
| 141 | ] | |
| 142 | }, | |
| 143 | { | |
| 144 | "cell_type": "markdown", | |
| 145 | "metadata": {}, | |
| 146 | "source": [ | |
| 147 | "## Controlling the level of detail" | |
| 100 | 148 | ] |
| 101 | 149 | }, |
| 102 | 150 | { |
| 115 | 163 | "metadata": {}, |
| 116 | 164 | "outputs": [], |
| 117 | 165 | "source": [ |
| 118 | "ax = df.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 119 | "ctx.add_basemap(ax, zoom=12)" | |
| 166 | "ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 167 | "cx.add_basemap(ax, zoom=12)" | |
| 168 | ] | |
| 169 | }, | |
| 170 | { | |
| 171 | "cell_type": "markdown", | |
| 172 | "metadata": {}, | |
| 173 | "source": [ | |
| 174 | "## Choosing a different style" | |
| 120 | 175 | ] |
| 121 | 176 | }, |
| 122 | 177 | { |
| 124 | 179 | "metadata": {}, |
| 125 | 180 | "source": [ |
| 126 | 181 | "By default, contextily uses the Stamen Terrain style. We can specify a\n", |
| 127 | "different style using ``ctx.providers``:\n", | |
| 182 | "different style using ``cx.providers``:\n", | |
| 128 | 183 | "\n" |
| 129 | 184 | ] |
| 130 | 185 | }, |
| 134 | 189 | "metadata": {}, |
| 135 | 190 | "outputs": [], |
| 136 | 191 | "source": [ |
| 137 | "ax = df.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 138 | "ctx.add_basemap(ax, url=ctx.providers.Stamen.TonerLite)\n", | |
| 192 | "ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 193 | "cx.add_basemap(ax, source=cx.providers.Stamen.TonerLite)\n", | |
| 139 | 194 | "ax.set_axis_off()" |
| 140 | 195 | ] |
| 141 | 196 | }, |
| 142 | 197 | { |
| 143 | "cell_type": "code", | |
| 144 | "execution_count": null, | |
| 145 | "metadata": {}, | |
| 146 | "outputs": [], | |
| 147 | "source": [] | |
| 198 | "cell_type": "markdown", | |
| 199 | "metadata": {}, | |
| 200 | "source": [ | |
| 201 | "## Adding labels as an overlay" | |
| 202 | ] | |
| 203 | }, | |
| 204 | { | |
| 205 | "cell_type": "markdown", | |
| 206 | "metadata": {}, | |
| 207 | "source": [ | |
| 208 | "Sometimes, when you plot data on a basemap, the data will obscure some important map elements, such as labels,\n", | |
| 209 | "that you would otherwise want to see unobscured. Some map tile providers offer multiple sets of partially\n", | |
| 210 | "transparent tiles to solve this, and `contextily` will do its best to auto-detect these transparent layers\n", | |
| 211 | "and put them on top." | |
| 212 | ] | |
| 213 | }, | |
| 214 | { | |
| 215 | "cell_type": "code", | |
| 216 | "execution_count": null, | |
| 217 | "metadata": {}, | |
| 218 | "outputs": [], | |
| 219 | "source": [ | |
| 220 | "ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 221 | "cx.add_basemap(ax, source=cx.providers.Stamen.TonerLite)\n", | |
| 222 | "cx.add_basemap(ax, source=cx.providers.Stamen.TonerLabels)" | |
| 223 | ] | |
| 224 | }, | |
| 225 | { | |
| 226 | "cell_type": "markdown", | |
| 227 | "metadata": {}, | |
| 228 | "source": [ | |
| 229 | "By splitting the layers like this, you can also independently manipulate the level of zoom on each layer,\n", | |
| 230 | "for example to make labels larger while still showing a lot of detail." | |
| 231 | ] | |
| 232 | }, | |
| 233 | { | |
| 234 | "cell_type": "code", | |
| 235 | "execution_count": null, | |
| 236 | "metadata": {}, | |
| 237 | "outputs": [], | |
| 238 | "source": [ | |
| 239 | "ax = df_wm.plot(figsize=(10, 10), alpha=0.5, edgecolor='k')\n", | |
| 240 | "cx.add_basemap(ax, source=cx.providers.Stamen.Watercolor, zoom=12)\n", | |
| 241 | "cx.add_basemap(ax, source=cx.providers.Stamen.TonerLabels, zoom=10)" | |
| 242 | ] | |
| 148 | 243 | } |
| 149 | 244 | ], |
| 150 | 245 | "metadata": { |
| 163 | 258 | "name": "python", |
| 164 | 259 | "nbconvert_exporter": "python", |
| 165 | 260 | "pygments_lexer": "ipython3", |
| 166 | "version": "3.7.6" | |
| 261 | "version": "3.9.1" | |
| 167 | 262 | } |
| 168 | 263 | }, |
| 169 | 264 | "nbformat": 4, |
| 3 | 3 | "cell_type": "markdown", |
| 4 | 4 | "metadata": {}, |
| 5 | 5 | "source": [ |
| 6 | "# Plotting with folium\n", | |
| 6 | "# Plotting with Folium\n", | |
| 7 | 7 | "\n", |
| 8 | 8 | "__What is Folium?__\n", |
| 9 | 9 | "\n", |
| 10 | "It builds on the data wrangling and a Python wrapper for leaflet.js. It makes it easy to visualize data in Python with minimal instructions.\n", | |
| 11 | "\n", | |
| 12 | "Folium expands on the data wrangling properties utilized in Python language and the mapping characteristics of the Leaflet.js library. Folium enables us to make an intuitive map and are is visualized in a Leaflet map after manipulating data in Python. Folium results are intuitive which makes this library helpful for dashboard building and easier to work with.\n", | |
| 13 | "\n", | |
| 14 | "Let's see the implementation of both GeoPandas and Folium:" | |
| 15 | ] | |
| 16 | }, | |
| 17 | { | |
| 18 | "cell_type": "code", | |
| 19 | "execution_count": null, | |
| 20 | "metadata": {}, | |
| 21 | "outputs": [], | |
| 22 | "source": [ | |
| 23 | "# Importing Libraries\n", | |
| 10 | "[Folium](https://python-visualization.github.io/folium/) builds on the data wrangling strengths of the Python ecosystem and the mapping strengths of the leaflet.js library. This allows you to manipulate your data in Geopandas and visualize it on a Leaflet map via Folium.\n", | |
| 11 | "\n", | |
| 12 | "In this example, we will first use Geopandas to load the geometries (volcano point data), and then create the Folium map with markers representing the different types of volcanoes." | |
| 13 | ] | |
| 14 | }, | |
| 15 | { | |
| 16 | "cell_type": "markdown", | |
| 17 | "metadata": {}, | |
| 18 | "source": [ | |
| 19 | "## Load geometries\n", | |
| 20 | "This example uses a freely available [volcano dataset](https://www.kaggle.com/texasdave/volcano-eruptions). We will be reading the csv file using pandas, and then convert the pandas `DataFrame` to a Geopandas `GeoDataFrame`." | |
| 21 | ] | |
| 22 | }, | |
| 23 | { | |
| 24 | "cell_type": "code", | |
| 25 | "execution_count": null, | |
| 26 | "metadata": {}, | |
| 27 | "outputs": [], | |
| 28 | "source": [ | |
| 29 | "# Import Libraries\n", | |
| 24 | 30 | "import pandas as pd\n", |
| 25 | 31 | "import geopandas\n", |
| 26 | 32 | "import folium\n", |
| 27 | "import matplotlib.pyplot as plt\n", | |
| 28 | "\n", | |
| 29 | "from shapely.geometry import Point" | |
| 33 | "import matplotlib.pyplot as plt" | |
| 30 | 34 | ] |
| 31 | 35 | }, |
| 32 | 36 | { |
| 36 | 40 | "outputs": [], |
| 37 | 41 | "source": [ |
| 38 | 42 | "df1 = pd.read_csv('volcano_data_2010.csv')\n", |
| 43 | "\n", | |
| 44 | "# Keep only relevant columns\n", | |
| 39 | 45 | "df = df1.loc[:, (\"Year\", \"Name\", \"Country\", \"Latitude\", \"Longitude\", \"Type\")]\n", |
| 40 | 46 | "df.info()" |
| 41 | 47 | ] |
| 46 | 52 | "metadata": {}, |
| 47 | 53 | "outputs": [], |
| 48 | 54 | "source": [ |
| 55 | "# Create point geometries\n", | |
| 49 | 56 | "geometry = geopandas.points_from_xy(df.Longitude, df.Latitude)\n", |
| 50 | 57 | "geo_df = geopandas.GeoDataFrame(df[['Year','Name','Country', 'Latitude', 'Longitude', 'Type']], geometry=geometry)\n", |
| 51 | 58 | "\n", |
| 82 | 89 | "cell_type": "markdown", |
| 83 | 90 | "metadata": {}, |
| 84 | 91 | "source": [ |
| 85 | "We will be using different icons to differentiate the types of Volcanoes using Folium.\n", | |
| 86 | "But before we start, we can see a few different tiles to choose from folium." | |
| 92 | "## Create Folium map\n", | |
| 93 | "Folium has a number of built-in tilesets from OpenStreetMap, Mapbox, and Stamen. For example:" | |
| 87 | 94 | ] |
| 88 | 95 | }, |
| 89 | 96 | { |
| 123 | 130 | "cell_type": "markdown", |
| 124 | 131 | "metadata": {}, |
| 125 | 132 | "source": [ |
| 126 | "We can use other tiles for the visualization, these are just a few examples.\n", | |
| 127 | "\n", | |
| 128 | "### Markers\n", | |
| 129 | "Now, let's look at different volcanoes on the map using different Markers to represent the volcanoes." | |
| 130 | ] | |
| 131 | }, | |
| 132 | { | |
| 133 | "cell_type": "code", | |
| 134 | "execution_count": null, | |
| 135 | "metadata": {}, | |
| 136 | "outputs": [], | |
| 137 | "source": [ | |
| 138 | "#use terrain map layer to actually see volcano terrain\n", | |
| 133 | "This example uses the Stamen Terrain map layer to visualize the volcano terrain." | |
| 134 | ] | |
| 135 | }, | |
| 136 | { | |
| 137 | "cell_type": "code", | |
| 138 | "execution_count": null, | |
| 139 | "metadata": {}, | |
| 140 | "outputs": [], | |
| 141 | "source": [ | |
| 142 | "# Use terrain map layer to see volcano terrain\n", | |
| 139 | 143 | "map = folium.Map(location = [4,10], tiles = \"Stamen Terrain\", zoom_start = 3)" |
| 140 | 144 | ] |
| 141 | 145 | }, |
| 142 | 146 | { |
| 143 | "cell_type": "code", | |
| 144 | "execution_count": null, | |
| 145 | "metadata": {}, | |
| 146 | "outputs": [], | |
| 147 | "source": [ | |
| 148 | "# insert multiple markers, iterate through list\n", | |
| 149 | "# add a different color marker associated with type of volcano\n", | |
| 150 | "\n", | |
| 147 | "cell_type": "markdown", | |
| 148 | "metadata": {}, | |
| 149 | "source": [ | |
| 150 | "### Add markers\n", | |
| 151 | "To represent the different types of volcanoes, you can create Folium markers and add them to your map." | |
| 152 | ] | |
| 153 | }, | |
| 154 | { | |
| 155 | "cell_type": "code", | |
| 156 | "execution_count": null, | |
| 157 | "metadata": {}, | |
| 158 | "outputs": [], | |
| 159 | "source": [ | |
| 160 | "# Create a geometry list from the GeoDataFrame\n", | |
| 151 | 161 | "geo_df_list = [[point.xy[1][0], point.xy[0][0]] for point in geo_df.geometry ]\n", |
| 152 | 162 | "\n", |
| 163 | "# Iterate through list and add a marker for each volcano, color-coded by its type.\n", | |
| 153 | 164 | "i = 0\n", |
| 154 | 165 | "for coordinates in geo_df_list:\n", |
| 155 | 166 | " #assign a color marker for the type of volcano, Strato being the most common\n", |
| 165 | 176 | " type_color = \"purple\"\n", |
| 166 | 177 | "\n", |
| 167 | 178 | "\n", |
| 168 | " #now place the markers with the popup labels and data\n", | |
| 179 | " # Place the markers with the popup labels and data\n", | |
| 169 | 180 | " map.add_child(folium.Marker(location = coordinates,\n", |
| 170 | 181 | " popup =\n", |
| 171 | 182 | " \"Year: \" + str(geo_df.Year[i]) + '<br>' +\n", |
| 190 | 201 | "cell_type": "markdown", |
| 191 | 202 | "metadata": {}, |
| 192 | 203 | "source": [ |
| 193 | "### Heatmaps\n", | |
| 194 | "\n", | |
| 195 | "Folium is well known for it's heatmap which create a heatmap layer. To plot a heat map in folium, one needs a list of Latitude, Longitude." | |
| 196 | ] | |
| 197 | }, | |
| 198 | { | |
| 199 | "cell_type": "code", | |
| 200 | "execution_count": null, | |
| 201 | "metadata": {}, | |
| 202 | "outputs": [], | |
| 203 | "source": [ | |
| 204 | "# In this example, with the hep of heat maps, we are able to perceive the density of volcanoes\n", | |
| 205 | "# which is more in some part of the world compared to others.\n", | |
| 204 | "## Folium Heatmaps\n", | |
| 205 | "\n", | |
| 206 | "Folium is well known for its heatmaps, which create a heatmap layer. To plot a heatmap in Folium, you need a list of latitudes and longitudes." | |
| 207 | ] | |
| 208 | }, | |
| 209 | { | |
| 210 | "cell_type": "code", | |
| 211 | "execution_count": null, | |
| 212 | "metadata": {}, | |
| 213 | "outputs": [], | |
| 214 | "source": [ | |
| 215 | "# This example uses heatmaps to visualize the density of volcanoes\n", | |
| 216 | "# which is more in some parts of the world compared to others.\n", | |
| 206 | 217 | "\n", |
| 207 | 218 | "from folium import plugins\n", |
| 208 | 219 | "\n", |
| 215 | 226 | "\n", |
| 216 | 227 | "map" |
| 217 | 228 | ] |
| 218 | }, | |
| 219 | { | |
| 220 | "cell_type": "code", | |
| 221 | "execution_count": null, | |
| 222 | "metadata": {}, | |
| 223 | "outputs": [], | |
| 224 | "source": [] | |
| 225 | 229 | } |
| 226 | 230 | ], |
| 227 | 231 | "metadata": { |
| 240 | 244 | "name": "python", |
| 241 | 245 | "nbconvert_exporter": "python", |
| 242 | 246 | "pygments_lexer": "ipython3", |
| 243 | "version": "3.7.6" | |
| 247 | "version": "3.9.1" | |
| 244 | 248 | } |
| 245 | 249 | }, |
| 246 | 250 | "nbformat": 4, |
| 3 | 3 | "cell_type": "markdown", |
| 4 | 4 | "metadata": {}, |
| 5 | 5 | "source": [ |
| 6 | "# An example of polygon plotting with folium \n", | |
| 7 | "We are going to demonstrate polygon plotting in this example with the help of folium" | |
| 6 | "# Plotting polygons with Folium\n", | |
| 7 | "This example demonstrates how to plot polygons on a Folium map." | |
| 8 | 8 | ] |
| 9 | 9 | }, |
| 10 | 10 | { |
| 22 | 22 | "cell_type": "markdown", |
| 23 | 23 | "metadata": {}, |
| 24 | 24 | "source": [ |
| 25 | "We make use of nybb dataset" | |
| 25 | "## Load geometries\n", | |
| 26 | "This example uses the nybb dataset, which contains polygons of New York boroughs." | |
| 26 | 27 | ] |
| 27 | 28 | }, |
| 28 | 29 | { |
| 61 | 62 | "cell_type": "markdown", |
| 62 | 63 | "metadata": {}, |
| 63 | 64 | "source": [ |
| 64 | "One thing to notice is that the values of the geometry do not directly represent the values of latitude of longitude in geographic coordinate system\n" | |
| 65 | ] | |
| 66 | }, | |
| 67 | { | |
| 68 | "cell_type": "code", | |
| 69 | "execution_count": null, | |
| 70 | "metadata": {}, | |
| 71 | "outputs": [], | |
| 72 | "source": [ | |
| 73 | "print(df.crs)" | |
| 74 | ] | |
| 75 | }, | |
| 76 | { | |
| 77 | "cell_type": "markdown", | |
| 78 | "metadata": {}, | |
| 79 | "source": [ | |
| 80 | "As folium(i.e. leaflet.js) by default takes input of values of latitude and longitude, we need to project the geometry first" | |
| 81 | ] | |
| 82 | }, | |
| 83 | { | |
| 84 | "cell_type": "code", | |
| 85 | "execution_count": null, | |
| 86 | "metadata": {}, | |
| 87 | "outputs": [], | |
| 88 | "source": [ | |
| 65 | "Notice that the values of the polygon geometries do not directly represent the values of latitude of longitude in a geographic coordinate system.\n", | |
| 66 | "To view the coordinate reference system of the geometry column, access the `crs` attribute:" | |
| 67 | ] | |
| 68 | }, | |
| 69 | { | |
| 70 | "cell_type": "code", | |
| 71 | "execution_count": null, | |
| 72 | "metadata": {}, | |
| 73 | "outputs": [], | |
| 74 | "source": [ | |
| 75 | "df.crs" | |
| 76 | ] | |
| 77 | }, | |
| 78 | { | |
| 79 | "cell_type": "markdown", | |
| 80 | "metadata": {}, | |
| 81 | "source": [ | |
| 82 | "The [epsg:2263](https://epsg.io/2263) crs is a projected coordinate reference system with linear units (ft in this case).\n", | |
| 83 | "As folium (i.e. leaflet.js) by default accepts values of latitude and longitude (angular units) as input, we need to project the geometry to a geographic coordinate system first." | |
| 84 | ] | |
| 85 | }, | |
| 86 | { | |
| 87 | "cell_type": "code", | |
| 88 | "execution_count": null, | |
| 89 | "metadata": {}, | |
| 90 | "outputs": [], | |
| 91 | "source": [ | |
| 92 | "# Use WGS 84 (epsg:4326) as the geographic coordinate system\n", | |
| 89 | 93 | "df = df.to_crs(epsg=4326)\n", |
| 90 | 94 | "print(df.crs)\n", |
| 91 | 95 | "df.head()" |
| 105 | 109 | "cell_type": "markdown", |
| 106 | 110 | "metadata": {}, |
| 107 | 111 | "source": [ |
| 108 | "Initialize folium map object" | |
| 112 | "## Create Folium map" | |
| 109 | 113 | ] |
| 110 | 114 | }, |
| 111 | 115 | { |
| 122 | 126 | "cell_type": "markdown", |
| 123 | 127 | "metadata": {}, |
| 124 | 128 | "source": [ |
| 125 | "Overlay the boundaries of boroughs on map with borough name as popup" | |
| 129 | "### Add polygons to map\n", | |
| 130 | "Overlay the boundaries of boroughs on map with borough name as popup:" | |
| 126 | 131 | ] |
| 127 | 132 | }, |
| 128 | 133 | { |
| 132 | 137 | "outputs": [], |
| 133 | 138 | "source": [ |
| 134 | 139 | "for _, r in df.iterrows():\n", |
| 135 | " #without simplifying the representation of each borough, the map might not be displayed \n", | |
| 136 | " #sim_geo = gpd.GeoSeries(r['geometry'])\n", | |
| 140 | " # Without simplifying the representation of each borough,\n", | |
| 141 | " # the map might not be displayed \n", | |
| 137 | 142 | " sim_geo = gpd.GeoSeries(r['geometry']).simplify(tolerance=0.001)\n", |
| 138 | 143 | " geo_j = sim_geo.to_json()\n", |
| 139 | 144 | " geo_j = folium.GeoJson(data=geo_j,\n", |
| 147 | 152 | "cell_type": "markdown", |
| 148 | 153 | "metadata": {}, |
| 149 | 154 | "source": [ |
| 150 | "Add marker showing the area and length of each borough" | |
| 151 | ] | |
| 152 | }, | |
| 153 | { | |
| 154 | "cell_type": "code", | |
| 155 | "execution_count": null, | |
| 156 | "metadata": {}, | |
| 157 | "outputs": [], | |
| 158 | "source": [ | |
| 159 | "df['lat'] = df.centroid.y\n", | |
| 160 | "df['lon'] = df.centroid.x\n", | |
| 155 | "### Add centroid markers\n", | |
| 156 | "In order to properly compute geometric properties, in this case centroids, of the geometries, we need to project the data to a projected coordinate system." | |
| 157 | ] | |
| 158 | }, | |
| 159 | { | |
| 160 | "cell_type": "code", | |
| 161 | "execution_count": null, | |
| 162 | "metadata": {}, | |
| 163 | "outputs": [], | |
| 164 | "source": [ | |
| 165 | "# Project to NAD83 projected crs\n", | |
| 166 | "df = df.to_crs(epsg=2263)\n", | |
| 167 | "\n", | |
| 168 | "# Access the centroid attribute of each polygon\n", | |
| 169 | "df['centroid'] = df.centroid" | |
| 170 | ] | |
| 171 | }, | |
| 172 | { | |
| 173 | "cell_type": "markdown", | |
| 174 | "metadata": {}, | |
| 175 | "source": [ | |
| 176 | "Since we're again adding a new geometry to the Folium map, we need to project the geometry back to a geographic coordinate system with latitude and longitude values." | |
| 177 | ] | |
| 178 | }, | |
| 179 | { | |
| 180 | "cell_type": "code", | |
| 181 | "execution_count": null, | |
| 182 | "metadata": {}, | |
| 183 | "outputs": [], | |
| 184 | "source": [ | |
| 185 | "# Project to WGS84 geographic crs\n", | |
| 186 | "\n", | |
| 187 | "# geometry (active) column\n", | |
| 188 | "df = df.to_crs(epsg=4326)\n", | |
| 189 | "\n", | |
| 190 | "# Centroid column\n", | |
| 191 | "df['centroid'] = df['centroid'].to_crs(epsg=4326)\n", | |
| 192 | "\n", | |
| 161 | 193 | "df.head()" |
| 162 | 194 | ] |
| 163 | 195 | }, |
| 168 | 200 | "outputs": [], |
| 169 | 201 | "source": [ |
| 170 | 202 | "for _, r in df.iterrows():\n", |
| 171 | " folium.Marker(location=[r['lat'], r['lon']], popup='length: {} <br> area: {}'.format(r['Shape_Leng'], r['Shape_Area'])).add_to(m)\n", | |
| 172 | " \n", | |
| 203 | " lat = r['centroid'].y\n", | |
| 204 | " lon = r['centroid'].x\n", | |
| 205 | " folium.Marker(location=[lat, lon],\n", | |
| 206 | " popup='length: {} <br> area: {}'.format(r['Shape_Leng'], r['Shape_Area'])).add_to(m)\n", | |
| 207 | "\n", | |
| 173 | 208 | "m" |
| 174 | 209 | ] |
| 175 | }, | |
| 176 | { | |
| 177 | "cell_type": "code", | |
| 178 | "execution_count": null, | |
| 179 | "metadata": {}, | |
| 180 | "outputs": [], | |
| 181 | "source": [] | |
| 182 | 210 | } |
| 183 | 211 | ], |
| 184 | 212 | "metadata": { |
| 213 | "kernelspec": { | |
| 214 | "display_name": "Python 3", | |
| 215 | "language": "python", | |
| 216 | "name": "python3" | |
| 217 | }, | |
| 185 | 218 | "language_info": { |
| 186 | 219 | "codemirror_mode": { |
| 187 | 220 | "name": "ipython", |
| 192 | 225 | "name": "python", |
| 193 | 226 | "nbconvert_exporter": "python", |
| 194 | 227 | "pygments_lexer": "ipython3", |
| 195 | "version": "3.8.5" | |
| 228 | "version": "3.9.1" | |
| 196 | 229 | } |
| 197 | 230 | }, |
| 198 | 231 | "nbformat": 4, |
Binary diff not shown
| 1 | 1 | "cells": [ |
| 2 | 2 | { |
| 3 | 3 | "cell_type": "markdown", |
| 4 | "metadata": {}, | |
| 5 | 4 | "source": [ |
| 6 | 5 | "# Spatial Joins\n", |
| 7 | 6 | "\n", |
| 12 | 11 | "A common use case might be a spatial join between a point layer and a polygon layer where you want to retain the point geometries and grab the attributes of the intersecting polygons.\n", |
| 13 | 12 | "\n", |
| 14 | 13 | "" |
| 15 | ] | |
| 16 | }, | |
| 17 | { | |
| 18 | "cell_type": "markdown", | |
| 19 | "metadata": {}, | |
| 14 | ], | |
| 15 | "metadata": {} | |
| 16 | }, | |
| 17 | { | |
| 18 | "cell_type": "markdown", | |
| 20 | 19 | "source": [ |
| 21 | 20 | "\n", |
| 22 | 21 | "## Types of spatial joins\n", |
| 84 | 83 | " 0101000000F0D88AA0E1A4EEBF7052F7E5B115E9BF | 2 | 20\n", |
| 85 | 84 | "(4 rows) \n", |
| 86 | 85 | "```" |
| 87 | ] | |
| 88 | }, | |
| 89 | { | |
| 90 | "cell_type": "markdown", | |
| 91 | "metadata": {}, | |
| 86 | ], | |
| 87 | "metadata": {} | |
| 88 | }, | |
| 89 | { | |
| 90 | "cell_type": "markdown", | |
| 92 | 91 | "source": [ |
| 93 | 92 | "## Spatial Joins between two GeoDataFrames\n", |
| 94 | 93 | "\n", |
| 95 | 94 | "Let's take a look at how we'd implement these using `GeoPandas`. First, load up the NYC test data into `GeoDataFrames`:" |
| 96 | ] | |
| 97 | }, | |
| 98 | { | |
| 99 | "cell_type": "code", | |
| 100 | "execution_count": null, | |
| 101 | "metadata": {}, | |
| 102 | "outputs": [], | |
| 95 | ], | |
| 96 | "metadata": {} | |
| 97 | }, | |
| 98 | { | |
| 99 | "cell_type": "code", | |
| 100 | "execution_count": null, | |
| 103 | 101 | "source": [ |
| 104 | 102 | "%matplotlib inline\n", |
| 105 | 103 | "from shapely.geometry import Point\n", |
| 106 | 104 | "from geopandas import datasets, GeoDataFrame, read_file\n", |
| 107 | "from geopandas.tools import overlay\n", | |
| 108 | 105 | "\n", |
| 109 | 106 | "# NYC Boros\n", |
| 110 | 107 | "zippath = datasets.get_path('nybb')\n", |
| 120 | 117 | "\n", |
| 121 | 118 | "# Make sure they're using the same projection reference\n", |
| 122 | 119 | "pointdf.crs = polydf.crs" |
| 123 | ] | |
| 124 | }, | |
| 125 | { | |
| 126 | "cell_type": "code", | |
| 127 | "execution_count": null, | |
| 128 | "metadata": {}, | |
| 129 | "outputs": [], | |
| 120 | ], | |
| 121 | "outputs": [], | |
| 122 | "metadata": {} | |
| 123 | }, | |
| 124 | { | |
| 125 | "cell_type": "code", | |
| 126 | "execution_count": null, | |
| 130 | 127 | "source": [ |
| 131 | 128 | "pointdf" |
| 132 | ] | |
| 133 | }, | |
| 134 | { | |
| 135 | "cell_type": "code", | |
| 136 | "execution_count": null, | |
| 137 | "metadata": {}, | |
| 138 | "outputs": [], | |
| 129 | ], | |
| 130 | "outputs": [], | |
| 131 | "metadata": {} | |
| 132 | }, | |
| 133 | { | |
| 134 | "cell_type": "code", | |
| 135 | "execution_count": null, | |
| 139 | 136 | "source": [ |
| 140 | 137 | "polydf" |
| 141 | ] | |
| 142 | }, | |
| 143 | { | |
| 144 | "cell_type": "code", | |
| 145 | "execution_count": null, | |
| 146 | "metadata": {}, | |
| 147 | "outputs": [], | |
| 138 | ], | |
| 139 | "outputs": [], | |
| 140 | "metadata": {} | |
| 141 | }, | |
| 142 | { | |
| 143 | "cell_type": "code", | |
| 144 | "execution_count": null, | |
| 148 | 145 | "source": [ |
| 149 | 146 | "pointdf.plot()" |
| 150 | ] | |
| 151 | }, | |
| 152 | { | |
| 153 | "cell_type": "code", | |
| 154 | "execution_count": null, | |
| 155 | "metadata": {}, | |
| 156 | "outputs": [], | |
| 147 | ], | |
| 148 | "outputs": [], | |
| 149 | "metadata": {} | |
| 150 | }, | |
| 151 | { | |
| 152 | "cell_type": "code", | |
| 153 | "execution_count": null, | |
| 157 | 154 | "source": [ |
| 158 | 155 | "polydf.plot()" |
| 159 | ] | |
| 160 | }, | |
| 161 | { | |
| 162 | "cell_type": "markdown", | |
| 163 | "metadata": {}, | |
| 156 | ], | |
| 157 | "outputs": [], | |
| 158 | "metadata": {} | |
| 159 | }, | |
| 160 | { | |
| 161 | "cell_type": "markdown", | |
| 164 | 162 | "source": [ |
| 165 | 163 | "## Joins" |
| 166 | ] | |
| 167 | }, | |
| 168 | { | |
| 169 | "cell_type": "code", | |
| 170 | "execution_count": null, | |
| 171 | "metadata": {}, | |
| 172 | "outputs": [], | |
| 173 | "source": [ | |
| 174 | "from geopandas.tools import sjoin\n", | |
| 175 | "join_left_df = sjoin(pointdf, polydf, how=\"left\")\n", | |
| 164 | ], | |
| 165 | "metadata": {} | |
| 166 | }, | |
| 167 | { | |
| 168 | "cell_type": "code", | |
| 169 | "execution_count": null, | |
| 170 | "source": [ | |
| 171 | "join_left_df = pointdf.sjoin(polydf, how=\"left\")\n", | |
| 176 | 172 | "join_left_df\n", |
| 177 | 173 | "# Note the NaNs where the point did not intersect a boro" |
| 178 | ] | |
| 179 | }, | |
| 180 | { | |
| 181 | "cell_type": "code", | |
| 182 | "execution_count": null, | |
| 183 | "metadata": {}, | |
| 184 | "outputs": [], | |
| 185 | "source": [ | |
| 186 | "join_right_df = sjoin(pointdf, polydf, how=\"right\")\n", | |
| 174 | ], | |
| 175 | "outputs": [], | |
| 176 | "metadata": {} | |
| 177 | }, | |
| 178 | { | |
| 179 | "cell_type": "code", | |
| 180 | "execution_count": null, | |
| 181 | "source": [ | |
| 182 | "join_right_df = pointdf.sjoin(polydf, how=\"right\")\n", | |
| 187 | 183 | "join_right_df\n", |
| 188 | 184 | "# Note Staten Island is repeated" |
| 189 | ] | |
| 190 | }, | |
| 191 | { | |
| 192 | "cell_type": "code", | |
| 193 | "execution_count": null, | |
| 194 | "metadata": {}, | |
| 195 | "outputs": [], | |
| 196 | "source": [ | |
| 197 | "join_inner_df = sjoin(pointdf, polydf, how=\"inner\")\n", | |
| 185 | ], | |
| 186 | "outputs": [], | |
| 187 | "metadata": {} | |
| 188 | }, | |
| 189 | { | |
| 190 | "cell_type": "code", | |
| 191 | "execution_count": null, | |
| 192 | "source": [ | |
| 193 | "join_inner_df = pointdf.sjoin(polydf, how=\"inner\")\n", | |
| 198 | 194 | "join_inner_df\n", |
| 199 | 195 | "# Note the lack of NaNs; dropped anything that didn't intersect" |
| 200 | ] | |
| 201 | }, | |
| 202 | { | |
| 203 | "cell_type": "markdown", | |
| 204 | "metadata": {}, | |
| 196 | ], | |
| 197 | "outputs": [], | |
| 198 | "metadata": {} | |
| 199 | }, | |
| 200 | { | |
| 201 | "cell_type": "markdown", | |
| 205 | 202 | "source": [ |
| 206 | 203 | "We're not limited to using the `intersection` binary predicate. Any of the `Shapely` geometry methods that return a Boolean can be used by specifying the `op` kwarg." |
| 207 | ] | |
| 208 | }, | |
| 209 | { | |
| 210 | "cell_type": "code", | |
| 211 | "execution_count": null, | |
| 212 | "metadata": {}, | |
| 213 | "outputs": [], | |
| 214 | "source": [ | |
| 215 | "sjoin(pointdf, polydf, how=\"left\", op=\"within\")" | |
| 216 | ] | |
| 204 | ], | |
| 205 | "metadata": {} | |
| 206 | }, | |
| 207 | { | |
| 208 | "cell_type": "code", | |
| 209 | "execution_count": null, | |
| 210 | "source": [ | |
| 211 | "pointdf.sjoin(polydf, how=\"left\", op=\"within\")" | |
| 212 | ], | |
| 213 | "outputs": [], | |
| 214 | "metadata": {} | |
| 217 | 215 | } |
| 218 | 216 | ], |
| 219 | 217 | "metadata": { |
| 237 | 235 | }, |
| 238 | 236 | "nbformat": 4, |
| 239 | 237 | "nbformat_minor": 4 |
| 240 | } | |
| 238 | }⏎ | |
| 137 | 137 | Required dependencies: |
| 138 | 138 | |
| 139 | 139 | - `numpy`_ |
| 140 | - `pandas`_ (version 0.24 or later) | |
| 140 | - `pandas`_ (version 0.25 or later) | |
| 141 | 141 | - `shapely`_ (interface to `GEOS`_) |
| 142 | 142 | - `fiona`_ (interface to `GDAL`_) |
| 143 | 143 | - `pyproj`_ (interface to `PROJ`_; version 2.2.0 or later) |
| 153 | 153 | |
| 154 | 154 | For plotting, these additional packages may be used: |
| 155 | 155 | |
| 156 | - `matplotlib`_ (>= 2.2.0) | |
| 157 | - `mapclassify`_ (>= 2.2.0) | |
| 156 | - `matplotlib`_ (>= 3.1.0) | |
| 157 | - `mapclassify`_ (>= 2.4.0) | |
| 158 | 158 | |
| 159 | 159 | |
| 160 | 160 | Using the optional PyGEOS dependency |
| 1 | 1 | "cells": [ |
| 2 | 2 | { |
| 3 | 3 | "cell_type": "markdown", |
| 4 | "metadata": {}, | |
| 4 | "metadata": { | |
| 5 | "tags": [] | |
| 6 | }, | |
| 5 | 7 | "source": [ |
| 6 | 8 | "# Introduction to GeoPandas\n", |
| 7 | 9 | "\n", |
| 8 | "This quick tutorial provides an introduction to the key concepts of GeoPandas. In a few minutes, we'll describe the basics which allow you to start your projects.\n", | |
| 10 | "This quick tutorial introduces the key concepts and basic features of GeoPandas to help you get started with your projects.\n", | |
| 9 | 11 | "\n", |
| 10 | 12 | "## Concepts\n", |
| 11 | 13 | "\n", |
| 12 | "GeoPandas, as the name suggests, extends popular data science library [pandas](https://pandas.pydata.org) by adding support for geospatial data. If you are not familiar with `pandas`, we recommend taking a quick look at its [Getting started documentation](https://pandas.pydata.org/docs/getting_started/index.html#getting-started) before proceeding.\n", | |
| 13 | "\n", | |
| 14 | "The core data structure in GeoPandas is `geopandas.GeoDataFrame`, a subclass of `pandas.DataFrame` able to store geometry columns and perform spatial operations. Geometries are handled by `geopandas.GeoSeries`, a subclass of `pandas.Series`. Therefore, your `GeoDataFrame` is a combination of `Series` with your data (numerical, boolean, text etc.) and `GeoSeries` with geometries (points, polygons etc.). You can have as many columns with geometries as you wish, there's no limit typical for desktop GIS software.\n", | |
| 14 | "GeoPandas, as the name suggests, extends the popular data science library [pandas](https://pandas.pydata.org) by adding support for geospatial data. If you are not familiar with `pandas`, we recommend taking a quick look at its [Getting started documentation](https://pandas.pydata.org/docs/getting_started/index.html#getting-started) before proceeding.\n", | |
| 15 | "\n", | |
| 16 | "The core data structure in GeoPandas is the `geopandas.GeoDataFrame`, a subclass of `pandas.DataFrame`, that can store geometry columns and perform spatial operations. The `geopandas.GeoSeries`, a subclass of `pandas.Series`, handles the geometries. Therefore, your `GeoDataFrame` is a combination of `pandas.Series`, with traditional data (numerical, boolean, text etc.), and `geopandas.GeoSeries`, with geometries (points, polygons etc.). You can have as many columns with geometries as you wish; there's no limit typical for desktop GIS software.\n", | |
| 15 | 17 | "\n", |
| 16 | 18 | "\n", |
| 17 | 19 | "\n", |
| 18 | "Each `GeoSeries` can contain any geometry type (we can even mix them within a single array) and has a `GeoSeries.crs` attribute, which stores information on the projection (CRS stands for Coordinate Reference System). Therefore, each `GeoSeries` in a `GeoDataFrame` can be in a different projection, allowing you to have, for example, multiple versions of the same geometry, just in a different CRS.\n", | |
| 19 | "\n", | |
| 20 | "One `GeoSeries` within a `GeoDataFrame` is seen as the _active_ geometry, which means that all geometric operations applied to a `GeoDataFrame` use the specified column.\n", | |
| 20 | "Each `GeoSeries` can contain any geometry type (you can even mix them within a single array) and has a `GeoSeries.crs` attribute, which stores information about the projection (CRS stands for Coordinate Reference System). Therefore, each `GeoSeries` in a `GeoDataFrame` can be in a different projection, allowing you to have, for example, multiple versions (different projections) of the same geometry.\n", | |
| 21 | "\n", | |
| 22 | "Only one `GeoSeries` in a `GeoDataFrame` is considered the _active_ geometry, which means that all geometric operations applied to a `GeoDataFrame` operate on this _active_ column.\n", | |
| 21 | 23 | "\n", |
| 22 | 24 | "\n", |
| 23 | 25 | "<div class=\"alert alert-info\">\n", |
| 27 | 29 | "</div>\n", |
| 28 | 30 | "\n", |
| 29 | 31 | "\n", |
| 30 | "Let's see how this works in practice.\n", | |
| 32 | "Let's see how some of these concepts work in practice.\n", | |
| 31 | 33 | "\n", |
| 32 | 34 | "## Reading and writing files\n", |
| 33 | 35 | "\n", |
| 34 | 36 | "First, we need to read some data.\n", |
| 35 | 37 | "\n", |
| 36 | "### Read files\n", | |
| 37 | "\n", | |
| 38 | "Assuming we have a file containing both data and geometry (e.g. GeoPackage, GeoJSON, Shapefile), we can easily read it using `geopandas.read_file` function, which automatically detects filetype and creates a `GeoDataFrame`. In this example, we'll use the `\"nybb\"` dataset, a map of New York boroughs which is part of GeoPandas installation. Therefore we need to get the path to the actual file. With your file, you specify a path as a string (`\"my_data/my_file.geojson\"`)." | |
| 38 | "### Reading files\n", | |
| 39 | "\n", | |
| 40 | "Assuming you have a file containing both data and geometry (e.g. GeoPackage, GeoJSON, Shapefile), you can read it using `geopandas.read_file()`, which automatically detects the filetype and creates a `GeoDataFrame`. This tutorial uses the `\"nybb\"` dataset, a map of New York boroughs, which is part of the GeoPandas installation. Therefore, we use `geopandas.datasets.get_path()` to retrieve the path to the dataset." | |
| 39 | 41 | ] |
| 40 | 42 | }, |
| 41 | 43 | { |
| 54 | 56 | }, |
| 55 | 57 | { |
| 56 | 58 | "cell_type": "markdown", |
| 57 | "metadata": {}, | |
| 58 | "source": [ | |
| 59 | "### Write files\n", | |
| 60 | "\n", | |
| 61 | "Writing a `GeoDataFrame` back to file is similarly simple, using `GeoDataFrame.to_file`. The default file format is Shapefile, but you can specify your own using `driver` keyword." | |
| 59 | "metadata": { | |
| 60 | "tags": [] | |
| 61 | }, | |
| 62 | "source": [ | |
| 63 | "### Writing files\n", | |
| 64 | "\n", | |
| 65 | "To write a `GeoDataFrame` back to file use `GeoDataFrame.to_file()`. The default file format is Shapefile, but you can specify your own with the `driver` keyword." | |
| 62 | 66 | ] |
| 63 | 67 | }, |
| 64 | 68 | { |
| 72 | 76 | }, |
| 73 | 77 | { |
| 74 | 78 | "cell_type": "markdown", |
| 75 | "metadata": {}, | |
| 79 | "metadata": { | |
| 80 | "tags": [] | |
| 81 | }, | |
| 76 | 82 | "source": [ |
| 77 | 83 | "<div class=\"alert alert-info\">\n", |
| 78 | 84 | "User Guide\n", |
| 82 | 88 | "\n", |
| 83 | 89 | "\n", |
| 84 | 90 | "\n", |
| 85 | "## Simple methods\n", | |
| 91 | "## Simple accessors and methods\n", | |
| 86 | 92 | "\n", |
| 87 | 93 | "Now we have our `GeoDataFrame` and can start working with its geometry. \n", |
| 88 | 94 | "\n", |
| 89 | "Since we have only one geometry column read from the file, it is automatically seen as the active geometry and methods used on `GeoDataFrame` will be applied to the `\"geometry\"` column.\n", | |
| 95 | "Since there was only one geometry column in the New York Boroughs dataset, this column automatically becomes the _active_ geometry and spatial methods used on the `GeoDataFrame` will be applied to the `\"geometry\"` column.\n", | |
| 90 | 96 | "\n", |
| 91 | 97 | "### Measuring area\n", |
| 92 | 98 | "\n", |
| 93 | "To measure the area of each polygon (or MultiPolygon in this specific case), we can use `GeoDataFrame.area` attribute, which returns a `pandas.Series`. Note that `GeoDataFrame.area` is just `GeoSeries.area` applied to an active geometry column.\n", | |
| 94 | "\n", | |
| 95 | "But first, we set the names of boroughs as an index, to make the results easier to read." | |
| 99 | "To measure the area of each polygon (or MultiPolygon in this specific case), access the `GeoDataFrame.area` attribute, which returns a `pandas.Series`. Note that `GeoDataFrame.area` is just `GeoSeries.area` applied to the _active_ geometry column.\n", | |
| 100 | "\n", | |
| 101 | "But first, to make the results easier to read, set the names of the boroughs as the index:" | |
| 96 | 102 | ] |
| 97 | 103 | }, |
| 98 | 104 | { |
| 120 | 126 | "source": [ |
| 121 | 127 | "### Getting polygon boundary and centroid\n", |
| 122 | 128 | "\n", |
| 123 | "To get just the boundary of each polygon (LineString), we can call `GeoDataFrame.boundary`." | |
| 129 | "To get the boundary of each polygon (LineString), access the `GeoDataFrame.boundary`:" | |
| 124 | 130 | ] |
| 125 | 131 | }, |
| 126 | 132 | { |
| 158 | 164 | "source": [ |
| 159 | 165 | "### Measuring distance\n", |
| 160 | 166 | "\n", |
| 161 | "We can also measure how far is each centroid from the first one." | |
| 167 | "We can also measure how far each centroid is from the first centroid location." | |
| 162 | 168 | ] |
| 163 | 169 | }, |
| 164 | 170 | { |
| 176 | 182 | "cell_type": "markdown", |
| 177 | 183 | "metadata": {}, |
| 178 | 184 | "source": [ |
| 179 | "It's still a DataFrame, so we have all the pandas functionality available to use on the geospatial dataset, and to do data manipulations with the attributes and geometry information together.\n", | |
| 180 | "\n", | |
| 181 | "For example, we can calculate average of the distance measured above (by accessing the `'distance'` column, and calling the `mean()` method on it):" | |
| 185 | "Note that `geopandas.GeoDataFrame` is a subclass of `pandas.DataFrame`, so we have all the pandas functionality available to use on the geospatial dataset — we can even perform data manipulations with the attributes and geometry information together.\n", | |
| 186 | "\n", | |
| 187 | "For example, to calculate the average of the distances measured above, access the 'distance' column and call the mean() method on it:" | |
| 182 | 188 | ] |
| 183 | 189 | }, |
| 184 | 190 | { |
| 196 | 202 | "source": [ |
| 197 | 203 | "## Making maps\n", |
| 198 | 204 | "\n", |
| 199 | "GeoPandas can also plot maps, so we can check how our geometries look like in space. The key method here is `GeoDataFrame.plot()`. In the example below, we plot the `\"area\"` we measured earlier using the active geometry column. We also want to show a legend (`legend=True`)." | |
| 205 | "GeoPandas can also plot maps, so we can check how the geometries appear in space. To plot the active geometry, call `GeoDataFrame.plot()`. To color code by another column, pass in that column as the first argument. In the example below, we plot the active geometry column and color code by the `\"area\"` column. We also want to show a legend (`legend=True`)." | |
| 200 | 206 | ] |
| 201 | 207 | }, |
| 202 | 208 | { |
| 206 | 212 | "outputs": [], |
| 207 | 213 | "source": [ |
| 208 | 214 | "gdf.plot(\"area\", legend=True)" |
| 215 | ] | |
| 216 | }, | |
| 217 | { | |
| 218 | "cell_type": "markdown", | |
| 219 | "metadata": {}, | |
| 220 | "source": [ | |
| 221 | "You can also explore your data interactively using `GeoDataFrame.explore()`, which behaves in the same way `plot()` does but returns an interactive map instead." | |
| 222 | ] | |
| 223 | }, | |
| 224 | { | |
| 225 | "cell_type": "code", | |
| 226 | "execution_count": null, | |
| 227 | "metadata": {}, | |
| 228 | "outputs": [], | |
| 229 | "source": [ | |
| 230 | "gdf.explore(\"area\", legend=False)" | |
| 209 | 231 | ] |
| 210 | 232 | }, |
| 211 | 233 | { |
| 274 | 296 | "\n", |
| 275 | 297 | "### Convex hull\n", |
| 276 | 298 | "\n", |
| 277 | "If we are interested in the convex hull of our polygons, we can call `GeoDataFrame.convex_hull`." | |
| 299 | "If we are interested in the convex hull of our polygons, we can access `GeoDataFrame.convex_hull`." | |
| 278 | 300 | ] |
| 279 | 301 | }, |
| 280 | 302 | { |
| 417 | 439 | { |
| 418 | 440 | "cell_type": "code", |
| 419 | 441 | "execution_count": null, |
| 420 | "metadata": {}, | |
| 442 | "metadata": { | |
| 443 | "tags": [] | |
| 444 | }, | |
| 421 | 445 | "outputs": [], |
| 422 | 446 | "source": [ |
| 423 | 447 | "gdf = gdf.set_geometry(\"buffered_centroid\")\n", |
| 431 | 455 | "source": [ |
| 432 | 456 | "## Projections\n", |
| 433 | 457 | "\n", |
| 434 | "Each `GeoSeries` has the Coordinate Reference System (CRS) accessible as `GeoSeries.crs`. CRS tells GeoPandas where the coordinates of geometries are located on the Earth. In some cases, CRS is geographic, which means that coordinates are in latitude and longitude. In those cases, its CRS is WGS84, with the authority code `EPSG:4326`. Let's see the projection of our NY boroughs `GeoDataFrame`." | |
| 458 | "Each `GeoSeries` has its Coordinate Reference System (CRS) accessible at `GeoSeries.crs`. The CRS tells GeoPandas where the coordinates of the geometries are located on the earth's surface. In some cases, the CRS is geographic, which means that the coordinates are in latitude and longitude. In those cases, its CRS is WGS84, with the authority code `EPSG:4326`. Let's see the projection of our NY boroughs `GeoDataFrame`." | |
| 435 | 459 | ] |
| 436 | 460 | }, |
| 437 | 461 | { |
| 474 | 498 | "cell_type": "markdown", |
| 475 | 499 | "metadata": {}, |
| 476 | 500 | "source": [ |
| 477 | "Notice the difference in coordinates along the axes of the plot. Where we had 120 000 - 280 000 (feet) before, we have 40.5 - 40.9 (degrees) now. In this case, `boroughs_4326` has a `\"geometry\"` column in WGS84 but all the other (with centroids etc.) remains in the original CRS.\n", | |
| 501 | "Notice the difference in coordinates along the axes of the plot. Where we had 120 000 - 280 000 (feet) before, we now have 40.5 - 40.9 (degrees). In this case, `boroughs_4326` has a `\"geometry\"` column in WGS84 but all the other (with centroids etc.) remain in the original CRS.\n", | |
| 478 | 502 | "\n", |
| 479 | 503 | "<div class=\"alert alert-warning\">\n", |
| 480 | 504 | "Warning\n", |
| 481 | 505 | " \n", |
| 482 | "For operations that rely on distance or area, you always need to use projected CRS (in meters, feet, kilometers etc.) not a geographic one. GeoPandas operations are planar, and degrees reflect the position on a sphere. Therefore the results may not be correct. For example, the result of `gdf.area.sum()` (projected CRS) is 8 429 911 572 ft<sup>2</sup> but the result of `boroughs_4326.area.sum()` (geographic CRS) is 0.083.\n", | |
| 506 | "For operations that rely on distance or area, you always need to use a projected CRS (in meters, feet, kilometers etc.) not a geographic one (in degrees). GeoPandas operations are planar, whereas degrees reflect the position on a sphere. Therefore, spatial operations using degrees may not yield correct results. For example, the result of `gdf.area.sum()` (projected CRS) is 8 429 911 572 ft<sup>2</sup> but the result of `boroughs_4326.area.sum()` (geographic CRS) is 0.083.\n", | |
| 483 | 507 | "</div>\n", |
| 484 | 508 | "\n", |
| 485 | 509 | "<div class=\"alert alert-info\">\n", |
| 490 | 514 | "\n", |
| 491 | 515 | "## What next?\n", |
| 492 | 516 | "\n", |
| 493 | "With GeoPandas we can do much more that this, from [aggregations](../docs/user_guide/aggregation_with_dissolve.rst), to [spatial joins](../docs/user_guide/mergingdata.rst), [geocoding](../docs/user_guide/geocoding.rst) and [much more](../gallery/index.rst).\n", | |
| 494 | "\n", | |
| 495 | "Head to the [User Guide](../docs/user_guide.rst) for to learn more about different functionality of GeoPandas, to the [Examples](../gallery/index.rst) to see how it can be used or the the [API reference](../docs/reference.rst) for the details." | |
| 496 | ] | |
| 497 | }, | |
| 498 | { | |
| 499 | "cell_type": "code", | |
| 500 | "execution_count": null, | |
| 501 | "metadata": {}, | |
| 502 | "outputs": [], | |
| 503 | "source": [] | |
| 517 | "With GeoPandas we can do much more than what has been introduced so far, from [aggregations](../docs/user_guide/aggregation_with_dissolve.rst), to [spatial joins](../docs/user_guide/mergingdata.rst), to [geocoding](../docs/user_guide/geocoding.rst), and [much more](../gallery/index.rst).\n", | |
| 518 | "\n", | |
| 519 | "Head over to the [User Guide](../docs/user_guide.rst) to learn more about the different features of GeoPandas, the [Examples](../gallery/index.rst) to see how they can be used, or to the [API reference](../docs/reference.rst) for the details." | |
| 520 | ] | |
| 504 | 521 | } |
| 505 | 522 | ], |
| 506 | 523 | "metadata": { |
| 524 | "kernelspec": { | |
| 525 | "display_name": "Python 3", | |
| 526 | "language": "python", | |
| 527 | "name": "python3" | |
| 528 | }, | |
| 507 | 529 | "language_info": { |
| 508 | 530 | "codemirror_mode": { |
| 509 | 531 | "name": "ipython", |
| 514 | 536 | "name": "python", |
| 515 | 537 | "nbconvert_exporter": "python", |
| 516 | 538 | "pygments_lexer": "ipython3", |
| 517 | "version": "3.7.6" | |
| 539 | "version": "3.9.2" | |
| 518 | 540 | } |
| 519 | 541 | }, |
| 520 | 542 | "nbformat": 4, |
| 13 | 13 | |
| 14 | 14 | ## Installation |
| 15 | 15 | |
| 16 | GeoPandas is written in pure Python, but has several dependecies written in C | |
| 16 | GeoPandas is written in pure Python, but has several dependencies written in C | |
| 17 | 17 | ([GEOS](https://geos.osgeo.org), [GDAL](https://www.gdal.org/), [PROJ](https://proj.org/)). Those base C libraries can sometimes be a challenge to |
| 18 | 18 | install. Therefore, we advise you to closely follow the recommendations below to avoid |
| 19 | 19 | installation problems. |
| 3 | 3 | dependencies: |
| 4 | 4 | # required |
| 5 | 5 | - fiona>=1.8 |
| 6 | - pandas>=0.24 | |
| 6 | - pandas>=0.25 | |
| 7 | 7 | - pyproj>=2.2.0 |
| 8 | 8 | - shapely>=1.6 |
| 9 | 9 |
| 7 | 7 | from geopandas.io.arrow import _read_parquet as read_parquet # noqa |
| 8 | 8 | from geopandas.io.arrow import _read_feather as read_feather # noqa |
| 9 | 9 | from geopandas.io.sql import _read_postgis as read_postgis # noqa |
| 10 | from geopandas.tools import sjoin # noqa | |
| 10 | from geopandas.tools import sjoin, sjoin_nearest # noqa | |
| 11 | 11 | from geopandas.tools import overlay # noqa |
| 12 | 12 | from geopandas.tools._show_versions import show_versions # noqa |
| 13 | 13 | from geopandas.tools import clip # noqa |
| 3 | 3 | import os |
| 4 | 4 | import warnings |
| 5 | 5 | |
| 6 | import numpy as np | |
| 6 | 7 | import pandas as pd |
| 7 | 8 | import pyproj |
| 8 | 9 | import shapely |
| 13 | 14 | # pandas compat |
| 14 | 15 | # ----------------------------------------------------------------------------- |
| 15 | 16 | |
| 16 | PANDAS_GE_025 = str(pd.__version__) >= LooseVersion("0.25.0") | |
| 17 | 17 | PANDAS_GE_10 = str(pd.__version__) >= LooseVersion("1.0.0") |
| 18 | 18 | PANDAS_GE_11 = str(pd.__version__) >= LooseVersion("1.1.0") |
| 19 | 19 | PANDAS_GE_115 = str(pd.__version__) >= LooseVersion("1.1.5") |
| 37 | 37 | PYGEOS_SHAPELY_COMPAT = None |
| 38 | 38 | |
| 39 | 39 | PYGEOS_GE_09 = None |
| 40 | PYGEOS_GE_010 = None | |
| 41 | ||
| 42 | INSTALL_PYGEOS_ERROR = "To use PyGEOS within GeoPandas, you need to install PyGEOS: \ | |
| 43 | 'conda install pygeos' or 'pip install pygeos'" | |
| 40 | 44 | |
| 41 | 45 | try: |
| 42 | 46 | import pygeos # noqa |
| 45 | 49 | if str(pygeos.__version__) >= LooseVersion("0.8"): |
| 46 | 50 | HAS_PYGEOS = True |
| 47 | 51 | PYGEOS_GE_09 = str(pygeos.__version__) >= LooseVersion("0.9") |
| 52 | PYGEOS_GE_010 = str(pygeos.__version__) >= LooseVersion("0.10") | |
| 48 | 53 | else: |
| 49 | 54 | warnings.warn( |
| 50 | 55 | "The installed version of PyGEOS is too old ({0} installed, 0.8 required)," |
| 88 | 93 | # validate the pygeos version |
| 89 | 94 | if not str(pygeos.__version__) >= LooseVersion("0.8"): |
| 90 | 95 | raise ImportError( |
| 91 | "PyGEOS >= 0.6 is required, version {0} is installed".format( | |
| 96 | "PyGEOS >= 0.8 is required, version {0} is installed".format( | |
| 92 | 97 | pygeos.__version__ |
| 93 | 98 | ) |
| 94 | 99 | ) |
| 114 | 119 | PYGEOS_SHAPELY_COMPAT = True |
| 115 | 120 | |
| 116 | 121 | except ImportError: |
| 117 | raise ImportError( | |
| 118 | "To use the PyGEOS speed-ups within GeoPandas, you need to install " | |
| 119 | "PyGEOS: 'conda install pygeos' or 'pip install pygeos'" | |
| 120 | ) | |
| 122 | raise ImportError(INSTALL_PYGEOS_ERROR) | |
| 121 | 123 | |
| 122 | 124 | |
| 123 | 125 | set_use_pygeos() |
| 143 | 145 | with warnings.catch_warnings(): |
| 144 | 146 | warnings.filterwarnings( |
| 145 | 147 | "ignore", "Iteration|The array interface|__len__", shapely_warning |
| 148 | ) | |
| 149 | yield | |
| 150 | ||
| 151 | ||
| 152 | elif (str(np.__version__) >= LooseVersion("1.21")) and not SHAPELY_GE_20: | |
| 153 | ||
| 154 | @contextlib.contextmanager | |
| 155 | def ignore_shapely2_warnings(): | |
| 156 | with warnings.catch_warnings(): | |
| 157 | # warning from numpy for existing Shapely releases (this is fixed | |
| 158 | # with Shapely 1.8) | |
| 159 | warnings.filterwarnings( | |
| 160 | "ignore", "An exception was ignored while fetching", DeprecationWarning | |
| 146 | 161 | ) |
| 147 | 162 | yield |
| 148 | 163 | |
| 209 | 224 | # ----------------------------------------------------------------------------- |
| 210 | 225 | |
| 211 | 226 | PYPROJ_LT_3 = LooseVersion(pyproj.__version__) < LooseVersion("3") |
| 227 | PYPROJ_GE_31 = LooseVersion(pyproj.__version__) >= LooseVersion("3.1") | |
| 15 | 15 | """Provide attribute-style access to configuration dict.""" |
| 16 | 16 | |
| 17 | 17 | def __init__(self, options): |
| 18 | super(Options, self).__setattr__("_options", options) | |
| 18 | super().__setattr__("_options", options) | |
| 19 | 19 | # populate with default values |
| 20 | 20 | config = {} |
| 21 | 21 | for key, option in options.items(): |
| 22 | 22 | config[key] = option.default_value |
| 23 | 23 | |
| 24 | super(Options, self).__setattr__("_config", config) | |
| 24 | super().__setattr__("_config", config) | |
| 25 | 25 | |
| 26 | 26 | def __setattr__(self, key, value): |
| 27 | 27 | # you can't set new keys |
| 58 | 58 | doc_text = "\n".join(textwrap.wrap(option.doc, width=70)) |
| 59 | 59 | else: |
| 60 | 60 | doc_text = u"No description available." |
| 61 | doc_text = indent(doc_text, prefix=" ") | |
| 61 | doc_text = textwrap.indent(doc_text, prefix=" ") | |
| 62 | 62 | description += doc_text + "\n" |
| 63 | 63 | space = "\n " |
| 64 | 64 | description = description.replace("\n", space) |
| 65 | 65 | return "{}({}{})".format(cls, space, description) |
| 66 | ||
| 67 | ||
| 68 | def indent(text, prefix, predicate=None): | |
| 69 | """ | |
| 70 | This is the python 3 textwrap.indent function, which is not available in | |
| 71 | python 2. | |
| 72 | """ | |
| 73 | if predicate is None: | |
| 74 | ||
| 75 | def predicate(line): | |
| 76 | return line.strip() | |
| 77 | ||
| 78 | def prefixed_lines(): | |
| 79 | for line in text.splitlines(True): | |
| 80 | yield (prefix + line if predicate(line) else line) | |
| 81 | ||
| 82 | return "".join(prefixed_lines()) | |
| 83 | 66 | |
| 84 | 67 | |
| 85 | 68 | def _validate_display_precision(value): |
| 0 | from textwrap import dedent | |
| 1 | from typing import Callable, Union | |
| 2 | ||
| 3 | ||
| 4 | # doc decorator function ported with modifications from Pandas | |
| 5 | # https://github.com/pandas-dev/pandas/blob/master/pandas/util/_decorators.py | |
| 6 | ||
| 7 | ||
| 8 | def doc(*docstrings: Union[str, Callable], **params) -> Callable: | |
| 9 | """ | |
| 10 | A decorator take docstring templates, concatenate them and perform string | |
| 11 | substitution on it. | |
| 12 | This decorator will add a variable "_docstring_components" to the wrapped | |
| 13 | callable to keep track the original docstring template for potential usage. | |
| 14 | If it should be consider as a template, it will be saved as a string. | |
| 15 | Otherwise, it will be saved as callable, and later user __doc__ and dedent | |
| 16 | to get docstring. | |
| 17 | ||
| 18 | Parameters | |
| 19 | ---------- | |
| 20 | *docstrings : str or callable | |
| 21 | The string / docstring / docstring template to be appended in order | |
| 22 | after default docstring under callable. | |
| 23 | **params | |
| 24 | The string which would be used to format docstring template. | |
| 25 | """ | |
| 26 | ||
| 27 | def decorator(decorated: Callable) -> Callable: | |
| 28 | # collecting docstring and docstring templates | |
| 29 | docstring_components: list[Union[str, Callable]] = [] | |
| 30 | if decorated.__doc__: | |
| 31 | docstring_components.append(dedent(decorated.__doc__)) | |
| 32 | ||
| 33 | for docstring in docstrings: | |
| 34 | if hasattr(docstring, "_docstring_components"): | |
| 35 | docstring_components.extend(docstring._docstring_components) | |
| 36 | elif isinstance(docstring, str) or docstring.__doc__: | |
| 37 | docstring_components.append(docstring) | |
| 38 | ||
| 39 | # formatting templates and concatenating docstring | |
| 40 | decorated.__doc__ = "".join( | |
| 41 | component.format(**params) | |
| 42 | if isinstance(component, str) | |
| 43 | else dedent(component.__doc__ or "") | |
| 44 | for component in docstring_components | |
| 45 | ) | |
| 46 | ||
| 47 | decorated._docstring_components = docstring_components | |
| 48 | return decorated | |
| 49 | ||
| 50 | return decorator |
| 6 | 6 | import warnings |
| 7 | 7 | |
| 8 | 8 | import numpy as np |
| 9 | import pandas as pd | |
| 9 | 10 | |
| 10 | 11 | import shapely.geometry |
| 11 | 12 | import shapely.geos |
| 43 | 44 | type_mapping, geometry_type_ids, geometry_type_values = None, None, None |
| 44 | 45 | |
| 45 | 46 | |
| 46 | def _isna(value): | |
| 47 | """ | |
| 48 | Check if scalar value is NA-like (None or np.nan). | |
| 47 | def isna(value): | |
| 48 | """ | |
| 49 | Check if scalar value is NA-like (None, np.nan or pd.NA). | |
| 49 | 50 | |
| 50 | 51 | Custom version that only works for scalars (returning True or False), |
| 51 | 52 | as `pd.isna` also works for array-like input returning a boolean array. |
| 53 | 54 | if value is None: |
| 54 | 55 | return True |
| 55 | 56 | elif isinstance(value, float) and np.isnan(value): |
| 57 | return True | |
| 58 | elif compat.PANDAS_GE_10 and value is pd.NA: | |
| 56 | 59 | return True |
| 57 | 60 | else: |
| 58 | 61 | return False |
| 126 | 129 | out.append(_shapely_to_pygeos(geom)) |
| 127 | 130 | else: |
| 128 | 131 | out.append(geom) |
| 129 | elif _isna(geom): | |
| 132 | elif isna(geom): | |
| 130 | 133 | out.append(None) |
| 131 | 134 | else: |
| 132 | 135 | raise TypeError("Input must be valid geometry objects: {0}".format(geom)) |
| 164 | 167 | out = [] |
| 165 | 168 | |
| 166 | 169 | for geom in data: |
| 167 | if geom is not None and len(geom): | |
| 168 | geom = shapely.wkb.loads(geom) | |
| 170 | if not isna(geom) and len(geom): | |
| 171 | geom = shapely.wkb.loads(geom, hex=isinstance(geom, str)) | |
| 169 | 172 | else: |
| 170 | 173 | geom = None |
| 171 | 174 | out.append(geom) |
| 199 | 202 | out = [] |
| 200 | 203 | |
| 201 | 204 | for geom in data: |
| 202 | if geom is not None and len(geom): | |
| 205 | if not isna(geom) and len(geom): | |
| 203 | 206 | if isinstance(geom, bytes): |
| 204 | 207 | geom = geom.decode("utf-8") |
| 205 | 208 | geom = shapely.wkt.loads(geom) |
| 246 | 249 | else: |
| 247 | 250 | out = _points_from_xy(x, y, z) |
| 248 | 251 | aout = np.empty(len(x), dtype=object) |
| 249 | aout[:] = out | |
| 252 | with compat.ignore_shapely2_warnings(): | |
| 253 | aout[:] = out | |
| 250 | 254 | return aout |
| 251 | 255 | |
| 252 | 256 | |
| 608 | 612 | "geometry types, None is returned." |
| 609 | 613 | ) |
| 610 | 614 | data = np.empty(len(data), dtype=object) |
| 611 | data[:] = inner_rings | |
| 615 | with compat.ignore_shapely2_warnings(): | |
| 616 | data[:] = inner_rings | |
| 612 | 617 | return data |
| 613 | 618 | |
| 614 | 619 | |
| 618 | 623 | else: |
| 619 | 624 | # method and not a property -> can't use _unary_geo |
| 620 | 625 | out = np.empty(len(data), dtype=object) |
| 621 | out[:] = [ | |
| 622 | geom.representative_point() if geom is not None else None for geom in data | |
| 623 | ] | |
| 626 | with compat.ignore_shapely2_warnings(): | |
| 627 | out[:] = [ | |
| 628 | geom.representative_point() if geom is not None else None | |
| 629 | for geom in data | |
| 630 | ] | |
| 624 | 631 | return out |
| 625 | 632 | |
| 626 | 633 | |
| 793 | 800 | |
| 794 | 801 | def interpolate(data, distance, normalized=False): |
| 795 | 802 | if compat.USE_PYGEOS: |
| 796 | return pygeos.line_interpolate_point(data, distance, normalize=normalized) | |
| 803 | try: | |
| 804 | return pygeos.line_interpolate_point(data, distance, normalized=normalized) | |
| 805 | except TypeError: # support for pygeos<0.9 | |
| 806 | return pygeos.line_interpolate_point(data, distance, normalize=normalized) | |
| 797 | 807 | else: |
| 798 | 808 | out = np.empty(len(data), dtype=object) |
| 799 | 809 | if isinstance(distance, np.ndarray): |
| 802 | 812 | "Length of distance sequence does not match " |
| 803 | 813 | "length of the GeoSeries" |
| 804 | 814 | ) |
| 815 | with compat.ignore_shapely2_warnings(): | |
| 816 | out[:] = [ | |
| 817 | geom.interpolate(dist, normalized=normalized) | |
| 818 | for geom, dist in zip(data, distance) | |
| 819 | ] | |
| 820 | return out | |
| 821 | ||
| 822 | with compat.ignore_shapely2_warnings(): | |
| 805 | 823 | out[:] = [ |
| 806 | geom.interpolate(dist, normalized=normalized) | |
| 807 | for geom, dist in zip(data, distance) | |
| 824 | geom.interpolate(distance, normalized=normalized) for geom in data | |
| 808 | 825 | ] |
| 809 | return out | |
| 810 | ||
| 811 | out[:] = [geom.interpolate(distance, normalized=normalized) for geom in data] | |
| 812 | 826 | return out |
| 813 | 827 | |
| 814 | 828 | |
| 857 | 871 | |
| 858 | 872 | def project(data, other, normalized=False): |
| 859 | 873 | if compat.USE_PYGEOS: |
| 860 | return pygeos.line_locate_point(data, other, normalize=normalized) | |
| 874 | try: | |
| 875 | return pygeos.line_locate_point(data, other, normalized=normalized) | |
| 876 | except TypeError: # support for pygeos<0.9 | |
| 877 | return pygeos.line_locate_point(data, other, normalize=normalized) | |
| 861 | 878 | else: |
| 862 | 879 | return _binary_op("project", data, other, normalized=normalized) |
| 863 | 880 | |
| 940 | 957 | result = np.empty(n, dtype=object) |
| 941 | 958 | for i in range(n): |
| 942 | 959 | geom = data[i] |
| 943 | if _isna(geom): | |
| 960 | if isna(geom): | |
| 944 | 961 | result[i] = geom |
| 945 | 962 | else: |
| 946 | 963 | result[i] = transform(func, geom) |
| 21 | 21 | # setup.py/versioneer.py will grep for the variable names, so they must |
| 22 | 22 | # each be defined on a line of their own. _version.py will just call |
| 23 | 23 | # get_keywords(). |
| 24 | git_refnames = " (tag: v0.9.0)" | |
| 25 | git_full = "ec4c6805d1182f846b9659345a5e66fa7c7afac7" | |
| 24 | git_refnames = " (HEAD -> master, tag: v0.10.0)" | |
| 25 | git_full = "0be92da324d6a83d2a65904cde5c983c433a1584" | |
| 26 | 26 | keywords = {"refnames": git_refnames, "full": git_full} |
| 27 | 27 | return keywords |
| 28 | 28 |
| 53 | 53 | |
| 54 | 54 | |
| 55 | 55 | register_extension_dtype(GeometryDtype) |
| 56 | ||
| 57 | ||
| 58 | def _isna(value): | |
| 59 | """ | |
| 60 | Check if scalar value is NA-like (None, np.nan or pd.NA). | |
| 61 | ||
| 62 | Custom version that only works for scalars (returning True or False), | |
| 63 | as `pd.isna` also works for array-like input returning a boolean array. | |
| 64 | """ | |
| 65 | if value is None: | |
| 66 | return True | |
| 67 | elif isinstance(value, float) and np.isnan(value): | |
| 68 | return True | |
| 69 | elif compat.PANDAS_GE_10 and value is pd.NA: | |
| 70 | return True | |
| 71 | else: | |
| 72 | return False | |
| 73 | 56 | |
| 74 | 57 | |
| 75 | 58 | def _check_crs(left, right, allow_none=False): |
| 397 | 380 | if isinstance(key, numbers.Integral): |
| 398 | 381 | raise ValueError("cannot set a single element with an array") |
| 399 | 382 | self.data[key] = value.data |
| 400 | elif isinstance(value, BaseGeometry) or _isna(value): | |
| 401 | if _isna(value): | |
| 383 | elif isinstance(value, BaseGeometry) or vectorized.isna(value): | |
| 384 | if vectorized.isna(value): | |
| 402 | 385 | # internally only use None as missing value indicator |
| 403 | 386 | # but accept others |
| 404 | 387 | value = None |
| 844 | 827 | raise RuntimeError("crs must be set to estimate UTM CRS.") |
| 845 | 828 | |
| 846 | 829 | minx, miny, maxx, maxy = self.total_bounds |
| 847 | # ensure using geographic coordinates | |
| 848 | if not self.crs.is_geographic: | |
| 849 | lon, lat = Transformer.from_crs( | |
| 850 | self.crs, "EPSG:4326", always_xy=True | |
| 851 | ).transform((minx, maxx, minx, maxx), (miny, miny, maxy, maxy)) | |
| 852 | x_center = np.mean(lon) | |
| 853 | y_center = np.mean(lat) | |
| 854 | else: | |
| 830 | if self.crs.is_geographic: | |
| 855 | 831 | x_center = np.mean([minx, maxx]) |
| 856 | 832 | y_center = np.mean([miny, maxy]) |
| 833 | # ensure using geographic coordinates | |
| 834 | else: | |
| 835 | transformer = Transformer.from_crs(self.crs, "EPSG:4326", always_xy=True) | |
| 836 | if compat.PYPROJ_GE_31: | |
| 837 | minx, miny, maxx, maxy = transformer.transform_bounds( | |
| 838 | minx, miny, maxx, maxy | |
| 839 | ) | |
| 840 | y_center = np.mean([miny, maxy]) | |
| 841 | # crossed the antimeridian | |
| 842 | if minx > maxx: | |
| 843 | # shift maxx from [-180,180] to [0,360] | |
| 844 | # so both numbers are positive for center calculation | |
| 845 | # Example: -175 to 185 | |
| 846 | maxx += 360 | |
| 847 | x_center = np.mean([minx, maxx]) | |
| 848 | # shift back to [-180,180] | |
| 849 | x_center = ((x_center + 180) % 360) - 180 | |
| 850 | else: | |
| 851 | x_center = np.mean([minx, maxx]) | |
| 852 | else: | |
| 853 | lon, lat = transformer.transform( | |
| 854 | (minx, maxx, minx, maxx), (miny, miny, maxy, maxy) | |
| 855 | ) | |
| 856 | x_center = np.mean(lon) | |
| 857 | y_center = np.mean(lat) | |
| 857 | 858 | |
| 858 | 859 | utm_crs_list = query_utm_crs_info( |
| 859 | 860 | datum_name=datum_name, |
| 967 | 968 | ) |
| 968 | 969 | # self.data[idx] = value |
| 969 | 970 | value_arr = np.empty(1, dtype=object) |
| 970 | value_arr[:] = [value] | |
| 971 | with compat.ignore_shapely2_warnings(): | |
| 972 | value_arr[:] = [value] | |
| 971 | 973 | self.data[idx] = value_arr |
| 972 | 974 | return self |
| 973 | 975 | |
| 1004 | 1006 | |
| 1005 | 1007 | if mask.any(): |
| 1006 | 1008 | # fill with value |
| 1007 | if _isna(value): | |
| 1009 | if vectorized.isna(value): | |
| 1008 | 1010 | value = None |
| 1009 | 1011 | elif not isinstance(value, BaseGeometry): |
| 1010 | 1012 | raise NotImplementedError( |
| 1046 | 1048 | pd_dtype = pd.api.types.pandas_dtype(dtype) |
| 1047 | 1049 | if isinstance(pd_dtype, pd.StringDtype): |
| 1048 | 1050 | # ensure to return a pandas string array instead of numpy array |
| 1049 | return pd.array(string_values, dtype="string") | |
| 1051 | return pd.array(string_values, dtype=pd_dtype) | |
| 1050 | 1052 | return string_values.astype(dtype, copy=False) |
| 1051 | 1053 | else: |
| 1052 | 1054 | return np.array(self, dtype=dtype, copy=copy) |
| 1059 | 1061 | return pygeos.is_missing(self.data) |
| 1060 | 1062 | else: |
| 1061 | 1063 | return np.array([g is None for g in self.data], dtype="bool") |
| 1064 | ||
| 1065 | def value_counts( | |
| 1066 | self, | |
| 1067 | dropna: bool = True, | |
| 1068 | ): | |
| 1069 | """ | |
| 1070 | Compute a histogram of the counts of non-null values. | |
| 1071 | ||
| 1072 | Parameters | |
| 1073 | ---------- | |
| 1074 | dropna : bool, default True | |
| 1075 | Don't include counts of NaN | |
| 1076 | ||
| 1077 | Returns | |
| 1078 | ------- | |
| 1079 | pd.Series | |
| 1080 | """ | |
| 1081 | ||
| 1082 | # note ExtensionArray usage of value_counts only specifies dropna, | |
| 1083 | # so sort, normalize and bins are not arguments | |
| 1084 | values = to_wkb(self) | |
| 1085 | from pandas import Series, Index | |
| 1086 | ||
| 1087 | result = Series(values).value_counts(dropna=dropna) | |
| 1088 | # value_counts converts None to nan, need to convert back for from_wkb to work | |
| 1089 | # note result.index already has object dtype, not geometry | |
| 1090 | # Can't use fillna(None) or Index.putmask, as this gets converted back to nan | |
| 1091 | # for object dtypes | |
| 1092 | result.index = Index( | |
| 1093 | from_wkb(np.where(result.index.isna(), None, result.index)) | |
| 1094 | ) | |
| 1095 | return result | |
| 1062 | 1096 | |
| 1063 | 1097 | def unique(self): |
| 1064 | 1098 | """Compute the ExtensionArray of unique values. |
| 1107 | 1141 | len(self) is returned, with all values filled with |
| 1108 | 1142 | ``self.dtype.na_value``. |
| 1109 | 1143 | """ |
| 1110 | shifted = super(GeometryArray, self).shift(periods, fill_value) | |
| 1144 | shifted = super().shift(periods, fill_value) | |
| 1111 | 1145 | shifted.crs = self.crs |
| 1112 | 1146 | return shifted |
| 1113 | 1147 | |
| 1223 | 1257 | |
| 1224 | 1258 | precision = geopandas.options.display_precision |
| 1225 | 1259 | if precision is None: |
| 1226 | # dummy heuristic based on 10 first geometries that should | |
| 1227 | # work in most cases | |
| 1228 | with warnings.catch_warnings(): | |
| 1229 | warnings.simplefilter("ignore", category=RuntimeWarning) | |
| 1230 | xmin, ymin, xmax, ymax = self[~self.isna()][:10].total_bounds | |
| 1231 | if ( | |
| 1232 | (-180 <= xmin <= 180) | |
| 1233 | and (-180 <= xmax <= 180) | |
| 1234 | and (-90 <= ymin <= 90) | |
| 1235 | and (-90 <= ymax <= 90) | |
| 1236 | ): | |
| 1237 | # geographic coordinates | |
| 1238 | precision = 5 | |
| 1260 | if self.crs: | |
| 1261 | if self.crs.is_projected: | |
| 1262 | precision = 3 | |
| 1263 | else: | |
| 1264 | precision = 5 | |
| 1239 | 1265 | else: |
| 1240 | # typically projected coordinates | |
| 1241 | # (in case of unit meter: mm precision) | |
| 1242 | precision = 3 | |
| 1266 | # fallback | |
| 1267 | # dummy heuristic based on 10 first geometries that should | |
| 1268 | # work in most cases | |
| 1269 | with warnings.catch_warnings(): | |
| 1270 | warnings.simplefilter("ignore", category=RuntimeWarning) | |
| 1271 | xmin, ymin, xmax, ymax = self[~self.isna()][:10].total_bounds | |
| 1272 | if ( | |
| 1273 | (-180 <= xmin <= 180) | |
| 1274 | and (-180 <= xmax <= 180) | |
| 1275 | and (-90 <= ymin <= 90) | |
| 1276 | and (-90 <= ymax <= 90) | |
| 1277 | ): | |
| 1278 | # geographic coordinates | |
| 1279 | precision = 5 | |
| 1280 | else: | |
| 1281 | # typically projected coordinates | |
| 1282 | # (in case of unit meter: mm precision) | |
| 1283 | precision = 3 | |
| 1243 | 1284 | return lambda geom: shapely.wkt.dumps(geom, rounding_precision=precision) |
| 1244 | 1285 | return repr |
| 1245 | 1286 | |
| 1318 | 1359 | """ |
| 1319 | 1360 | Return for `item in self`. |
| 1320 | 1361 | """ |
| 1321 | if _isna(item): | |
| 1362 | if vectorized.isna(item): | |
| 1322 | 1363 | if ( |
| 1323 | 1364 | item is self.dtype.na_value |
| 1324 | 1365 | or isinstance(item, self.dtype.type) |
| 5 | 5 | |
| 6 | 6 | from shapely.geometry import box |
| 7 | 7 | from shapely.geometry.base import BaseGeometry |
| 8 | from shapely.ops import cascaded_union | |
| 9 | 8 | |
| 10 | 9 | from .array import GeometryArray, GeometryDtype |
| 11 | 10 | |
| 698 | 697 | |
| 699 | 698 | @property |
| 700 | 699 | def cascaded_union(self): |
| 701 | """Deprecated: Return the unary_union of all geometries""" | |
| 702 | return cascaded_union(np.asarray(self.geometry.values)) | |
| 700 | """Deprecated: use `unary_union` instead""" | |
| 701 | warn( | |
| 702 | "The 'cascaded_union' attribute is deprecated, use 'unary_union' instead", | |
| 703 | FutureWarning, | |
| 704 | stacklevel=2, | |
| 705 | ) | |
| 706 | return self.geometry.values.unary_union() | |
| 703 | 707 | |
| 704 | 708 | @property |
| 705 | 709 | def unary_union(self): |
| 718 | 722 | |
| 719 | 723 | >>> union = s.unary_union |
| 720 | 724 | >>> print(union) |
| 721 | POLYGON ((0 0, 0 1, 0 2, 2 2, 2 0, 1 0, 0 0)) | |
| 725 | POLYGON ((0 1, 0 2, 2 2, 2 0, 1 0, 0 0, 0 1)) | |
| 722 | 726 | """ |
| 723 | 727 | return self.geometry.values.unary_union() |
| 724 | 728 | |
| 730 | 734 | """Returns a ``Series`` of ``dtype('bool')`` with value ``True`` for |
| 731 | 735 | each aligned geometry that contains `other`. |
| 732 | 736 | |
| 733 | An object is said to contain `other` if its `interior` contains the | |
| 734 | `boundary` and `interior` of the other object and their boundaries do | |
| 735 | not touch at all. | |
| 737 | An object is said to contain `other` if at least one point of `other` lies in | |
| 738 | the interior and no points of `other` lie in the exterior of the object. | |
| 739 | (Therefore, any given polygon does not contain its own boundary – there is not | |
| 740 | any point that lies in the interior.) | |
| 741 | If either object is empty, this operation returns ``False``. | |
| 736 | 742 | |
| 737 | 743 | This is the inverse of :meth:`within` in the sense that the expression |
| 738 | 744 | ``a.contains(b) == b.within(a)`` always evaluates to ``True``. |
| 745 | 751 | Parameters |
| 746 | 752 | ---------- |
| 747 | 753 | other : GeoSeries or geometric object |
| 748 | The GeoSeries (elementwise) or geometric object to test if is | |
| 749 | contained. | |
| 754 | The GeoSeries (elementwise) or geometric object to test if it | |
| 755 | is contained. | |
| 750 | 756 | align : bool (default True) |
| 751 | 757 | If True, automatically aligns GeoSeries based on their indices. |
| 752 | 758 | If False, the order of elements is preserved. |
| 1640 | 1646 | """Returns a ``Series`` of ``dtype('bool')`` with value ``True`` for |
| 1641 | 1647 | each aligned geometry that is within `other`. |
| 1642 | 1648 | |
| 1643 | An object is said to be within `other` if its `boundary` and `interior` | |
| 1644 | intersects only with the `interior` of the other (not its `boundary` or | |
| 1645 | `exterior`). | |
| 1649 | An object is said to be within `other` if at least one of its points is located | |
| 1650 | in the `interior` and no points are located in the `exterior` of the other. | |
| 1651 | If either object is empty, this operation returns ``False``. | |
| 1646 | 1652 | |
| 1647 | 1653 | This is the inverse of :meth:`contains` in the sense that the |
| 1648 | 1654 | expression ``a.within(b) == b.contains(a)`` always evaluates to |
| 1757 | 1763 | |
| 1758 | 1764 | An object A is said to cover another object B if no points of B lie |
| 1759 | 1765 | in the exterior of A. |
| 1766 | If either object is empty, this operation returns ``False``. | |
| 1760 | 1767 | |
| 1761 | 1768 | The operation works on a 1-to-1 row-wise manner: |
| 1762 | 1769 | |
| 2147 | 2154 | :align: center |
| 2148 | 2155 | |
| 2149 | 2156 | >>> s.difference(Polygon([(0, 0), (1, 1), (0, 1)])) |
| 2150 | 0 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2151 | 1 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2157 | 0 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2158 | 1 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2152 | 2159 | 2 LINESTRING (1.00000 1.00000, 2.00000 2.00000) |
| 2153 | 2160 | 3 MULTILINESTRING ((2.00000 0.00000, 1.00000 1.0... |
| 2154 | 2161 | 4 POINT EMPTY |
| 2164 | 2171 | |
| 2165 | 2172 | >>> s.difference(s2, align=True) |
| 2166 | 2173 | 0 None |
| 2167 | 1 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2174 | 1 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2168 | 2175 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2169 | 2176 | 3 LINESTRING EMPTY |
| 2170 | 2177 | 4 POINT (0.00000 1.00000) |
| 2172 | 2179 | dtype: geometry |
| 2173 | 2180 | |
| 2174 | 2181 | >>> s.difference(s2, align=False) |
| 2175 | 0 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2176 | 1 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2182 | 0 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2183 | 1 POLYGON ((0.00000 0.00000, 0.00000 2.00000, 1.... | |
| 2177 | 2184 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2178 | 2185 | 3 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2179 | 2186 | 4 POINT EMPTY |
| 2262 | 2269 | :align: center |
| 2263 | 2270 | |
| 2264 | 2271 | >>> s.symmetric_difference(Polygon([(0, 0), (1, 1), (0, 1)])) |
| 2265 | 0 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2266 | 1 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2267 | 2 GEOMETRYCOLLECTION (LINESTRING (1.00000 1.0000... | |
| 2268 | 3 GEOMETRYCOLLECTION (LINESTRING (2.00000 0.0000... | |
| 2269 | 4 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2272 | 0 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2273 | 1 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2274 | 2 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2275 | 3 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2276 | 4 POLYGON ((0.00000 1.00000, 1.00000 1.00000, 0.... | |
| 2270 | 2277 | dtype: geometry |
| 2271 | 2278 | |
| 2272 | 2279 | We can also check two GeoSeries against each other, row by row. |
| 2279 | 2286 | |
| 2280 | 2287 | >>> s.symmetric_difference(s2, align=True) |
| 2281 | 2288 | 0 None |
| 2282 | 1 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2289 | 1 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2283 | 2290 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2284 | 2291 | 3 LINESTRING EMPTY |
| 2285 | 2292 | 4 MULTIPOINT (0.00000 1.00000, 1.00000 1.00000) |
| 2287 | 2294 | dtype: geometry |
| 2288 | 2295 | |
| 2289 | 2296 | >>> s.symmetric_difference(s2, align=False) |
| 2290 | 0 POLYGON ((0.00000 1.00000, 0.00000 2.00000, 2.... | |
| 2291 | 1 GEOMETRYCOLLECTION (LINESTRING (1.00000 0.0000... | |
| 2297 | 0 POLYGON ((0.00000 2.00000, 2.00000 2.00000, 1.... | |
| 2298 | 1 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2292 | 2299 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2293 | 2300 | 3 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2294 | 2301 | 4 POINT EMPTY |
| 2374 | 2381 | :align: center |
| 2375 | 2382 | |
| 2376 | 2383 | >>> s.union(Polygon([(0, 0), (1, 1), (0, 1)])) |
| 2377 | 0 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2378 | 1 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2379 | 2 GEOMETRYCOLLECTION (LINESTRING (1.00000 1.0000... | |
| 2380 | 3 GEOMETRYCOLLECTION (LINESTRING (2.00000 0.0000... | |
| 2381 | 4 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2384 | 0 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 0.... | |
| 2385 | 1 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 0.... | |
| 2386 | 2 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2387 | 3 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2388 | 4 POLYGON ((0.00000 1.00000, 1.00000 1.00000, 0.... | |
| 2382 | 2389 | dtype: geometry |
| 2383 | 2390 | |
| 2384 | 2391 | We can also check two GeoSeries against each other, row by row. |
| 2391 | 2398 | |
| 2392 | 2399 | >>> s.union(s2, align=True) |
| 2393 | 2400 | 0 None |
| 2394 | 1 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2401 | 1 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 0.... | |
| 2395 | 2402 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2396 | 2403 | 3 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2397 | 2404 | 4 MULTIPOINT (0.00000 1.00000, 1.00000 1.00000) |
| 2399 | 2406 | dtype: geometry |
| 2400 | 2407 | |
| 2401 | 2408 | >>> s.union(s2, align=False) |
| 2402 | 0 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2403 | 1 GEOMETRYCOLLECTION (LINESTRING (1.00000 0.0000... | |
| 2409 | 0 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 0.... | |
| 2410 | 1 GEOMETRYCOLLECTION (POLYGON ((0.00000 0.00000,... | |
| 2404 | 2411 | 2 MULTILINESTRING ((0.00000 0.00000, 1.00000 1.0... |
| 2405 | 2412 | 3 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2406 | 2413 | 4 POINT (0.00000 1.00000) |
| 2487 | 2494 | :align: center |
| 2488 | 2495 | |
| 2489 | 2496 | >>> s.intersection(Polygon([(0, 0), (1, 1), (0, 1)])) |
| 2490 | 0 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2491 | 1 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2497 | 0 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2498 | 1 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2492 | 2499 | 2 LINESTRING (0.00000 0.00000, 1.00000 1.00000) |
| 2493 | 2500 | 3 POINT (1.00000 1.00000) |
| 2494 | 2501 | 4 POINT (0.00000 1.00000) |
| 2504 | 2511 | |
| 2505 | 2512 | >>> s.intersection(s2, align=True) |
| 2506 | 2513 | 0 None |
| 2507 | 1 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2514 | 1 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2508 | 2515 | 2 POINT (1.00000 1.00000) |
| 2509 | 2516 | 3 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2510 | 2517 | 4 POINT EMPTY |
| 2512 | 2519 | dtype: geometry |
| 2513 | 2520 | |
| 2514 | 2521 | >>> s.intersection(s2, align=False) |
| 2515 | 0 POLYGON ((1.00000 1.00000, 0.00000 0.00000, 0.... | |
| 2522 | 0 POLYGON ((0.00000 0.00000, 0.00000 1.00000, 1.... | |
| 2516 | 2523 | 1 LINESTRING (1.00000 1.00000, 1.00000 2.00000) |
| 2517 | 2524 | 2 POINT (1.00000 1.00000) |
| 2518 | 2525 | 3 POINT (1.00000 1.00000) |
| 2720 | 2727 | """Returns a ``GeoSeries`` containing a simplified representation of |
| 2721 | 2728 | each geometry. |
| 2722 | 2729 | |
| 2730 | The algorithm (Douglas-Peucker) recursively splits the original line | |
| 2731 | into smaller parts and connects these parts’ endpoints | |
| 2732 | by a straight line. Then, it removes all points whose distance | |
| 2733 | to the straight line is smaller than `tolerance`. It does not | |
| 2734 | move any points and it always preserves endpoints of | |
| 2735 | the original line or polygon. | |
| 2723 | 2736 | See http://shapely.readthedocs.io/en/latest/manual.html#object.simplify |
| 2724 | 2737 | for details |
| 2725 | 2738 | |
| 2726 | 2739 | Parameters |
| 2727 | 2740 | ---------- |
| 2728 | 2741 | tolerance : float |
| 2729 | All points in a simplified geometry will be no more than | |
| 2730 | `tolerance` distance from the original. | |
| 2742 | All parts of a simplified geometry will be no more than | |
| 2743 | `tolerance` distance from the original. It has the same units | |
| 2744 | as the coordinate reference system of the GeoSeries. | |
| 2745 | For example, using `tolerance=100` in a projected CRS with meters | |
| 2746 | as units means a distance of 100 meters in reality. | |
| 2731 | 2747 | preserve_topology: bool (default True) |
| 2732 | 2748 | False uses a quicker algorithm, but may produce self-intersecting |
| 2733 | 2749 | or otherwise invalid geometries. |
| 2891 | 2907 | |
| 2892 | 2908 | Examples |
| 2893 | 2909 | -------- |
| 2894 | >>> from shapely.geometry import Polygon, LineString, Point | |
| 2895 | >>> s = geopandas.GeoSeries( | |
| 2896 | ... [ | |
| 2897 | ... Polygon([(0, 0), (2, 2), (0, 2)]), | |
| 2910 | >>> from shapely.geometry import LineString, Point | |
| 2911 | >>> s = geopandas.GeoSeries( | |
| 2912 | ... [ | |
| 2913 | ... LineString([(0, 0), (2, 0), (0, 2)]), | |
| 2898 | 2914 | ... LineString([(0, 0), (2, 2)]), |
| 2899 | 2915 | ... LineString([(2, 0), (0, 2)]), |
| 2900 | 2916 | ... ], |
| 2909 | 2925 | ... ) |
| 2910 | 2926 | |
| 2911 | 2927 | >>> s |
| 2912 | 0 POLYGON ((0.00000 0.00000, 2.00000 2.00000, 0.... | |
| 2928 | 0 LINESTRING (0.00000 0.00000, 2.00000 0.00000, ... | |
| 2913 | 2929 | 1 LINESTRING (0.00000 0.00000, 2.00000 2.00000) |
| 2914 | 2930 | 2 LINESTRING (2.00000 0.00000, 0.00000 2.00000) |
| 2915 | 2931 | dtype: geometry |
| 2927 | 2943 | :align: center |
| 2928 | 2944 | |
| 2929 | 2945 | >>> s.project(Point(1, 0)) |
| 2930 | 0 -1.000000 | |
| 2946 | 0 1.000000 | |
| 2931 | 2947 | 1 0.707107 |
| 2932 | 2948 | 2 0.707107 |
| 2933 | 2949 | dtype: float64 |
| 2948 | 2964 | dtype: float64 |
| 2949 | 2965 | |
| 2950 | 2966 | >>> s.project(s2, align=False) |
| 2951 | 0 -1.000000 | |
| 2967 | 0 1.000000 | |
| 2952 | 2968 | 1 0.707107 |
| 2953 | 2969 | 2 0.707107 |
| 2954 | 2970 | dtype: float64 |
| 3300 | 3316 | # don't know how to handle step; should this raise? |
| 3301 | 3317 | if xs.step is not None or ys.step is not None: |
| 3302 | 3318 | warn("Ignoring step - full interval is used.") |
| 3303 | xmin, ymin, xmax, ymax = obj.total_bounds | |
| 3319 | if xs.start is None or xs.stop is None or ys.start is None or ys.stop is None: | |
| 3320 | xmin, ymin, xmax, ymax = obj.total_bounds | |
| 3304 | 3321 | bbox = box( |
| 3305 | 3322 | xs.start if xs.start is not None else xmin, |
| 3306 | 3323 | ys.start if ys.start is not None else ymin, |
| 0 | from statistics import mean | |
| 1 | ||
| 2 | import geopandas | |
| 3 | from shapely.geometry import LineString | |
| 4 | import numpy as np | |
| 5 | import pandas as pd | |
| 6 | ||
| 7 | _MAP_KWARGS = [ | |
| 8 | "location", | |
| 9 | "prefer_canvas", | |
| 10 | "no_touch", | |
| 11 | "disable_3d", | |
| 12 | "png_enabled", | |
| 13 | "zoom_control", | |
| 14 | "crs", | |
| 15 | "zoom_start", | |
| 16 | "left", | |
| 17 | "top", | |
| 18 | "position", | |
| 19 | "min_zoom", | |
| 20 | "max_zoom", | |
| 21 | "min_lat", | |
| 22 | "max_lat", | |
| 23 | "min_lon", | |
| 24 | "max_lon", | |
| 25 | "max_bounds", | |
| 26 | ] | |
| 27 | ||
| 28 | ||
| 29 | def _explore( | |
| 30 | df, | |
| 31 | column=None, | |
| 32 | cmap=None, | |
| 33 | color=None, | |
| 34 | m=None, | |
| 35 | tiles="OpenStreetMap", | |
| 36 | attr=None, | |
| 37 | tooltip=True, | |
| 38 | popup=False, | |
| 39 | highlight=True, | |
| 40 | categorical=False, | |
| 41 | legend=True, | |
| 42 | scheme=None, | |
| 43 | k=5, | |
| 44 | vmin=None, | |
| 45 | vmax=None, | |
| 46 | width="100%", | |
| 47 | height="100%", | |
| 48 | categories=None, | |
| 49 | classification_kwds=None, | |
| 50 | control_scale=True, | |
| 51 | marker_type=None, | |
| 52 | marker_kwds={}, | |
| 53 | style_kwds={}, | |
| 54 | highlight_kwds={}, | |
| 55 | missing_kwds={}, | |
| 56 | tooltip_kwds={}, | |
| 57 | popup_kwds={}, | |
| 58 | legend_kwds={}, | |
| 59 | **kwargs, | |
| 60 | ): | |
| 61 | """Interactive map based on GeoPandas and folium/leaflet.js | |
| 62 | ||
| 63 | Generate an interactive leaflet map based on :class:`~geopandas.GeoDataFrame` | |
| 64 | ||
| 65 | Parameters | |
| 66 | ---------- | |
| 67 | column : str, np.array, pd.Series (default None) | |
| 68 | The name of the dataframe column, :class:`numpy.array`, | |
| 69 | or :class:`pandas.Series` to be plotted. If :class:`numpy.array` or | |
| 70 | :class:`pandas.Series` are used then it must have same length as dataframe. | |
| 71 | cmap : str, matplotlib.Colormap, branca.colormap or function (default None) | |
| 72 | The name of a colormap recognized by ``matplotlib``, a list-like of colors, | |
| 73 | :class:`matplotlib.colors.Colormap`, a :class:`branca.colormap.ColorMap` or | |
| 74 | function that returns a named color or hex based on the column | |
| 75 | value, e.g.:: | |
| 76 | ||
| 77 | def my_colormap(value): # scalar value defined in 'column' | |
| 78 | if value > 1: | |
| 79 | return "green" | |
| 80 | return "red" | |
| 81 | ||
| 82 | color : str, array-like (default None) | |
| 83 | Named color or a list-like of colors (named or hex). | |
| 84 | m : folium.Map (default None) | |
| 85 | Existing map instance on which to draw the plot. | |
| 86 | tiles : str, xyzservices.TileProvider (default 'OpenStreetMap Mapnik') | |
| 87 | Map tileset to use. Can choose from the list supported by folium, query a | |
| 88 | :class:`xyzservices.TileProvider` by a name from ``xyzservices.providers``, | |
| 89 | pass :class:`xyzservices.TileProvider` object or pass custom XYZ URL. | |
| 90 | The current list of built-in providers (when ``xyzservices`` is not available): | |
| 91 | ||
| 92 | ``["OpenStreetMap", "Stamen Terrain", “Stamen Toner", “Stamen Watercolor" | |
| 93 | "CartoDB positron", “CartoDB dark_matter"]`` | |
| 94 | ||
| 95 | You can pass a custom tileset to Folium by passing a Leaflet-style URL | |
| 96 | to the tiles parameter: ``http://{s}.yourtiles.com/{z}/{x}/{y}.png``. | |
| 97 | Be sure to check their terms and conditions and to provide attribution with | |
| 98 | the ``attr`` keyword. | |
| 99 | attr : str (default None) | |
| 100 | Map tile attribution; only required if passing custom tile URL. | |
| 101 | tooltip : bool, str, int, list (default True) | |
| 102 | Display GeoDataFrame attributes when hovering over the object. | |
| 103 | ``True`` includes all columns. ``False`` removes tooltip. Pass string or list of | |
| 104 | strings to specify a column(s). Integer specifies first n columns to be | |
| 105 | included. Defaults to ``True``. | |
| 106 | popup : bool, str, int, list (default False) | |
| 107 | Input GeoDataFrame attributes for object displayed when clicking. | |
| 108 | ``True`` includes all columns. ``False`` removes popup. Pass string or list of | |
| 109 | strings to specify a column(s). Integer specifies first n columns to be | |
| 110 | included. Defaults to ``False``. | |
| 111 | highlight : bool (default True) | |
| 112 | Enable highlight functionality when hovering over a geometry. | |
| 113 | categorical : bool (default False) | |
| 114 | If ``False``, ``cmap`` will reflect numerical values of the | |
| 115 | column being plotted. For non-numerical columns, this | |
| 116 | will be set to True. | |
| 117 | legend : bool (default True) | |
| 118 | Plot a legend in choropleth plots. | |
| 119 | Ignored if no ``column`` is given. | |
| 120 | scheme : str (default None) | |
| 121 | Name of a choropleth classification scheme (requires ``mapclassify`` >= 2.4.0). | |
| 122 | A :func:`mapclassify.classify` will be used | |
| 123 | under the hood. Supported are all schemes provided by ``mapclassify`` (e.g. | |
| 124 | ``'BoxPlot'``, ``'EqualInterval'``, ``'FisherJenks'``, ``'FisherJenksSampled'``, | |
| 125 | ``'HeadTailBreaks'``, ``'JenksCaspall'``, ``'JenksCaspallForced'``, | |
| 126 | ``'JenksCaspallSampled'``, ``'MaxP'``, ``'MaximumBreaks'``, | |
| 127 | ``'NaturalBreaks'``, ``'Quantiles'``, ``'Percentiles'``, ``'StdMean'``, | |
| 128 | ``'UserDefined'``). Arguments can be passed in ``classification_kwds``. | |
| 129 | k : int (default 5) | |
| 130 | Number of classes | |
| 131 | vmin : None or float (default None) | |
| 132 | Minimum value of ``cmap``. If ``None``, the minimum data value | |
| 133 | in the column to be plotted is used. | |
| 134 | vmax : None or float (default None) | |
| 135 | Maximum value of ``cmap``. If ``None``, the maximum data value | |
| 136 | in the column to be plotted is used. | |
| 137 | width : pixel int or percentage string (default: '100%') | |
| 138 | Width of the folium :class:`~folium.folium.Map`. If the argument | |
| 139 | m is given explicitly, width is ignored. | |
| 140 | height : pixel int or percentage string (default: '100%') | |
| 141 | Height of the folium :class:`~folium.folium.Map`. If the argument | |
| 142 | m is given explicitly, height is ignored. | |
| 143 | categories : list-like | |
| 144 | Ordered list-like object of categories to be used for categorical plot. | |
| 145 | classification_kwds : dict (default None) | |
| 146 | Keyword arguments to pass to mapclassify | |
| 147 | control_scale : bool, (default True) | |
| 148 | Whether to add a control scale on the map. | |
| 149 | marker_type : str, folium.Circle, folium.CircleMarker, folium.Marker (default None) | |
| 150 | Allowed string options are ('marker', 'circle', 'circle_marker'). Defaults to | |
| 151 | folium.CircleMarker. | |
| 152 | marker_kwds: dict (default {}) | |
| 153 | Additional keywords to be passed to the selected ``marker_type``, e.g.: | |
| 154 | ||
| 155 | radius : float (default 2 for ``circle_marker`` and 50 for ``circle``)) | |
| 156 | Radius of the circle, in meters (for ``circle``) or pixels | |
| 157 | (for ``circle_marker``). | |
| 158 | fill : bool (default True) | |
| 159 | Whether to fill the ``circle`` or ``circle_marker`` with color. | |
| 160 | icon : folium.map.Icon | |
| 161 | the :class:`folium.map.Icon` object to use to render the marker. | |
| 162 | draggable : bool (default False) | |
| 163 | Set to True to be able to drag the marker around the map. | |
| 164 | ||
| 165 | style_kwds : dict (default {}) | |
| 166 | Additional style to be passed to folium ``style_function``: | |
| 167 | ||
| 168 | stroke : bool (default True) | |
| 169 | Whether to draw stroke along the path. Set it to ``False`` to | |
| 170 | disable borders on polygons or circles. | |
| 171 | color : str | |
| 172 | Stroke color | |
| 173 | weight : int | |
| 174 | Stroke width in pixels | |
| 175 | opacity : float (default 1.0) | |
| 176 | Stroke opacity | |
| 177 | fill : boolean (default True) | |
| 178 | Whether to fill the path with color. Set it to ``False`` to | |
| 179 | disable filling on polygons or circles. | |
| 180 | fillColor : str | |
| 181 | Fill color. Defaults to the value of the color option | |
| 182 | fillOpacity : float (default 0.5) | |
| 183 | Fill opacity. | |
| 184 | ||
| 185 | Plus all supported by :func:`folium.vector_layers.path_options`. See the | |
| 186 | documentation of :class:`folium.features.GeoJson` for details. | |
| 187 | ||
| 188 | highlight_kwds : dict (default {}) | |
| 189 | Style to be passed to folium highlight_function. Uses the same keywords | |
| 190 | as ``style_kwds``. When empty, defaults to ``{"fillOpacity": 0.75}``. | |
| 191 | tooltip_kwds : dict (default {}) | |
| 192 | Additional keywords to be passed to :class:`folium.features.GeoJsonTooltip`, | |
| 193 | e.g. ``aliases``, ``labels``, or ``sticky``. | |
| 194 | popup_kwds : dict (default {}) | |
| 195 | Additional keywords to be passed to :class:`folium.features.GeoJsonPopup`, | |
| 196 | e.g. ``aliases`` or ``labels``. | |
| 197 | legend_kwds : dict (default {}) | |
| 198 | Additional keywords to be passed to the legend. | |
| 199 | ||
| 200 | Currently supported customisation: | |
| 201 | ||
| 202 | caption : string | |
| 203 | Custom caption of the legend. Defaults to the column name. | |
| 204 | ||
| 205 | Additional accepted keywords when ``scheme`` is specified: | |
| 206 | ||
| 207 | colorbar : bool (default True) | |
| 208 | An option to control the style of the legend. If True, continuous | |
| 209 | colorbar will be used. If False, categorical legend will be used for bins. | |
| 210 | scale : bool (default True) | |
| 211 | Scale bins along the colorbar axis according to the bin edges (True) | |
| 212 | or use the equal length for each bin (False) | |
| 213 | fmt : string (default "{:.2f}") | |
| 214 | A formatting specification for the bin edges of the classes in the | |
| 215 | legend. For example, to have no decimals: ``{"fmt": "{:.0f}"}``. Applies | |
| 216 | if ``colorbar=False``. | |
| 217 | labels : list-like | |
| 218 | A list of legend labels to override the auto-generated labels. | |
| 219 | Needs to have the same number of elements as the number of | |
| 220 | classes (`k`). Applies if ``colorbar=False``. | |
| 221 | interval : boolean (default False) | |
| 222 | An option to control brackets from mapclassify legend. | |
| 223 | If True, open/closed interval brackets are shown in the legend. | |
| 224 | Applies if ``colorbar=False``. | |
| 225 | max_labels : int, default 10 | |
| 226 | Maximum number of colorbar tick labels (requires branca>=0.5.0) | |
| 227 | ||
| 228 | **kwargs : dict | |
| 229 | Additional options to be passed on to the folium object. | |
| 230 | ||
| 231 | Returns | |
| 232 | ------- | |
| 233 | m : folium.folium.Map | |
| 234 | folium :class:`~folium.folium.Map` instance | |
| 235 | ||
| 236 | Examples | |
| 237 | -------- | |
| 238 | >>> df = geopandas.read_file(geopandas.datasets.get_path("naturalearth_lowres")) | |
| 239 | >>> df.head(2) # doctest: +SKIP | |
| 240 | pop_est continent name iso_a3 \ | |
| 241 | gdp_md_est geometry | |
| 242 | 0 920938 Oceania Fiji FJI 8374.0 MULTIPOLY\ | |
| 243 | GON (((180.00000 -16.06713, 180.00000... | |
| 244 | 1 53950935 Africa Tanzania TZA 150600.0 POLYGON (\ | |
| 245 | (33.90371 -0.95000, 34.07262 -1.05982... | |
| 246 | ||
| 247 | >>> df.explore("pop_est", cmap="Blues") # doctest: +SKIP | |
| 248 | """ | |
| 249 | try: | |
| 250 | import branca as bc | |
| 251 | import folium | |
| 252 | import matplotlib.cm as cm | |
| 253 | import matplotlib.colors as colors | |
| 254 | import matplotlib.pyplot as plt | |
| 255 | from mapclassify import classify | |
| 256 | except (ImportError, ModuleNotFoundError): | |
| 257 | raise ImportError( | |
| 258 | "The 'folium', 'matplotlib' and 'mapclassify' packages are required for " | |
| 259 | "'explore()'. You can install them using " | |
| 260 | "'conda install -c conda-forge folium matplotlib mapclassify' " | |
| 261 | "or 'pip install folium matplotlib mapclassify'." | |
| 262 | ) | |
| 263 | ||
| 264 | # xyservices is an optional dependency | |
| 265 | try: | |
| 266 | import xyzservices | |
| 267 | ||
| 268 | HAS_XYZSERVICES = True | |
| 269 | except (ImportError, ModuleNotFoundError): | |
| 270 | HAS_XYZSERVICES = False | |
| 271 | ||
| 272 | gdf = df.copy() | |
| 273 | ||
| 274 | # convert LinearRing to LineString | |
| 275 | rings_mask = df.geom_type == "LinearRing" | |
| 276 | if rings_mask.any(): | |
| 277 | gdf.geometry[rings_mask] = gdf.geometry[rings_mask].apply( | |
| 278 | lambda g: LineString(g) | |
| 279 | ) | |
| 280 | ||
| 281 | if gdf.crs is None: | |
| 282 | kwargs["crs"] = "Simple" | |
| 283 | tiles = None | |
| 284 | elif not gdf.crs.equals(4326): | |
| 285 | gdf = gdf.to_crs(4326) | |
| 286 | ||
| 287 | # create folium.Map object | |
| 288 | if m is None: | |
| 289 | # Get bounds to specify location and map extent | |
| 290 | bounds = gdf.total_bounds | |
| 291 | location = kwargs.pop("location", None) | |
| 292 | if location is None: | |
| 293 | x = mean([bounds[0], bounds[2]]) | |
| 294 | y = mean([bounds[1], bounds[3]]) | |
| 295 | location = (y, x) | |
| 296 | if "zoom_start" in kwargs.keys(): | |
| 297 | fit = False | |
| 298 | else: | |
| 299 | fit = True | |
| 300 | else: | |
| 301 | fit = False | |
| 302 | ||
| 303 | # get a subset of kwargs to be passed to folium.Map | |
| 304 | map_kwds = {i: kwargs[i] for i in kwargs.keys() if i in _MAP_KWARGS} | |
| 305 | ||
| 306 | if HAS_XYZSERVICES: | |
| 307 | # match provider name string to xyzservices.TileProvider | |
| 308 | if isinstance(tiles, str): | |
| 309 | try: | |
| 310 | tiles = xyzservices.providers.query_name(tiles) | |
| 311 | except ValueError: | |
| 312 | pass | |
| 313 | ||
| 314 | if isinstance(tiles, xyzservices.TileProvider): | |
| 315 | attr = attr if attr else tiles.html_attribution | |
| 316 | map_kwds["min_zoom"] = tiles.get("min_zoom", 0) | |
| 317 | map_kwds["max_zoom"] = tiles.get("max_zoom", 18) | |
| 318 | tiles = tiles.build_url(scale_factor="{r}") | |
| 319 | ||
| 320 | m = folium.Map( | |
| 321 | location=location, | |
| 322 | control_scale=control_scale, | |
| 323 | tiles=tiles, | |
| 324 | attr=attr, | |
| 325 | width=width, | |
| 326 | height=height, | |
| 327 | **map_kwds, | |
| 328 | ) | |
| 329 | ||
| 330 | # fit bounds to get a proper zoom level | |
| 331 | if fit: | |
| 332 | m.fit_bounds([[bounds[1], bounds[0]], [bounds[3], bounds[2]]]) | |
| 333 | ||
| 334 | for map_kwd in _MAP_KWARGS: | |
| 335 | kwargs.pop(map_kwd, None) | |
| 336 | ||
| 337 | nan_idx = None | |
| 338 | ||
| 339 | if column is not None: | |
| 340 | if pd.api.types.is_list_like(column): | |
| 341 | if len(column) != gdf.shape[0]: | |
| 342 | raise ValueError( | |
| 343 | "The GeoDataFrame and given column have different number of rows." | |
| 344 | ) | |
| 345 | else: | |
| 346 | column_name = "__plottable_column" | |
| 347 | gdf[column_name] = column | |
| 348 | column = column_name | |
| 349 | elif pd.api.types.is_categorical_dtype(gdf[column]): | |
| 350 | if categories is not None: | |
| 351 | raise ValueError( | |
| 352 | "Cannot specify 'categories' when column has categorical dtype" | |
| 353 | ) | |
| 354 | categorical = True | |
| 355 | elif gdf[column].dtype is np.dtype("O") or categories: | |
| 356 | categorical = True | |
| 357 | ||
| 358 | nan_idx = pd.isna(gdf[column]) | |
| 359 | ||
| 360 | if categorical: | |
| 361 | cat = pd.Categorical(gdf[column][~nan_idx], categories=categories) | |
| 362 | N = len(cat.categories) | |
| 363 | cmap = cmap if cmap else "tab20" | |
| 364 | ||
| 365 | # colormap exists in matplotlib | |
| 366 | if cmap in plt.colormaps(): | |
| 367 | ||
| 368 | color = np.apply_along_axis( | |
| 369 | colors.to_hex, 1, cm.get_cmap(cmap, N)(cat.codes) | |
| 370 | ) | |
| 371 | legend_colors = np.apply_along_axis( | |
| 372 | colors.to_hex, 1, cm.get_cmap(cmap, N)(range(N)) | |
| 373 | ) | |
| 374 | ||
| 375 | # colormap is matplotlib.Colormap | |
| 376 | elif isinstance(cmap, colors.Colormap): | |
| 377 | color = np.apply_along_axis(colors.to_hex, 1, cmap(cat.codes)) | |
| 378 | legend_colors = np.apply_along_axis(colors.to_hex, 1, cmap(range(N))) | |
| 379 | ||
| 380 | # custom list of colors | |
| 381 | elif pd.api.types.is_list_like(cmap): | |
| 382 | if N > len(cmap): | |
| 383 | cmap = cmap * (N // len(cmap) + 1) | |
| 384 | color = np.take(cmap, cat.codes) | |
| 385 | legend_colors = np.take(cmap, range(N)) | |
| 386 | ||
| 387 | else: | |
| 388 | raise ValueError( | |
| 389 | "'cmap' is invalid. For categorical plots, pass either valid " | |
| 390 | "named matplotlib colormap or a list-like of colors." | |
| 391 | ) | |
| 392 | ||
| 393 | elif callable(cmap): | |
| 394 | # List of colors based on Branca colormaps or self-defined functions | |
| 395 | color = list(map(lambda x: cmap(x), df[column])) | |
| 396 | ||
| 397 | else: | |
| 398 | vmin = gdf[column].min() if not vmin else vmin | |
| 399 | vmax = gdf[column].max() if not vmax else vmax | |
| 400 | ||
| 401 | # get bins | |
| 402 | if scheme is not None: | |
| 403 | ||
| 404 | if classification_kwds is None: | |
| 405 | classification_kwds = {} | |
| 406 | if "k" not in classification_kwds: | |
| 407 | classification_kwds["k"] = k | |
| 408 | ||
| 409 | binning = classify( | |
| 410 | np.asarray(gdf[column][~nan_idx]), scheme, **classification_kwds | |
| 411 | ) | |
| 412 | color = np.apply_along_axis( | |
| 413 | colors.to_hex, 1, cm.get_cmap(cmap, k)(binning.yb) | |
| 414 | ) | |
| 415 | ||
| 416 | else: | |
| 417 | ||
| 418 | bins = np.linspace(vmin, vmax, 257)[1:] | |
| 419 | binning = classify( | |
| 420 | np.asarray(gdf[column][~nan_idx]), "UserDefined", bins=bins | |
| 421 | ) | |
| 422 | ||
| 423 | color = np.apply_along_axis( | |
| 424 | colors.to_hex, 1, cm.get_cmap(cmap, 256)(binning.yb) | |
| 425 | ) | |
| 426 | ||
| 427 | # set default style | |
| 428 | if "fillOpacity" not in style_kwds: | |
| 429 | style_kwds["fillOpacity"] = 0.5 | |
| 430 | if "weight" not in style_kwds: | |
| 431 | style_kwds["weight"] = 2 | |
| 432 | ||
| 433 | # specify color | |
| 434 | if color is not None: | |
| 435 | if ( | |
| 436 | isinstance(color, str) | |
| 437 | and isinstance(gdf, geopandas.GeoDataFrame) | |
| 438 | and color in gdf.columns | |
| 439 | ): # use existing column | |
| 440 | ||
| 441 | def _style_color(x): | |
| 442 | return { | |
| 443 | "fillColor": x["properties"][color], | |
| 444 | **style_kwds, | |
| 445 | } | |
| 446 | ||
| 447 | style_function = _style_color | |
| 448 | else: # assign new column | |
| 449 | if isinstance(gdf, geopandas.GeoSeries): | |
| 450 | gdf = geopandas.GeoDataFrame(geometry=gdf) | |
| 451 | ||
| 452 | if nan_idx is not None and nan_idx.any(): | |
| 453 | nan_color = missing_kwds.pop("color", None) | |
| 454 | ||
| 455 | gdf["__folium_color"] = nan_color | |
| 456 | gdf.loc[~nan_idx, "__folium_color"] = color | |
| 457 | else: | |
| 458 | gdf["__folium_color"] = color | |
| 459 | ||
| 460 | stroke_color = style_kwds.pop("color", None) | |
| 461 | if not stroke_color: | |
| 462 | ||
| 463 | def _style_column(x): | |
| 464 | return { | |
| 465 | "fillColor": x["properties"]["__folium_color"], | |
| 466 | "color": x["properties"]["__folium_color"], | |
| 467 | **style_kwds, | |
| 468 | } | |
| 469 | ||
| 470 | style_function = _style_column | |
| 471 | else: | |
| 472 | ||
| 473 | def _style_stroke(x): | |
| 474 | return { | |
| 475 | "fillColor": x["properties"]["__folium_color"], | |
| 476 | "color": stroke_color, | |
| 477 | **style_kwds, | |
| 478 | } | |
| 479 | ||
| 480 | style_function = _style_stroke | |
| 481 | else: # use folium default | |
| 482 | ||
| 483 | def _style_default(x): | |
| 484 | return {**style_kwds} | |
| 485 | ||
| 486 | style_function = _style_default | |
| 487 | ||
| 488 | if highlight: | |
| 489 | if "fillOpacity" not in highlight_kwds: | |
| 490 | highlight_kwds["fillOpacity"] = 0.75 | |
| 491 | ||
| 492 | def _style_highlight(x): | |
| 493 | return {**highlight_kwds} | |
| 494 | ||
| 495 | highlight_function = _style_highlight | |
| 496 | else: | |
| 497 | highlight_function = None | |
| 498 | ||
| 499 | # define default for points | |
| 500 | if marker_type is None: | |
| 501 | marker_type = "circle_marker" | |
| 502 | ||
| 503 | marker = marker_type | |
| 504 | if isinstance(marker_type, str): | |
| 505 | if marker_type == "marker": | |
| 506 | marker = folium.Marker(**marker_kwds) | |
| 507 | elif marker_type == "circle": | |
| 508 | marker = folium.Circle(**marker_kwds) | |
| 509 | elif marker_type == "circle_marker": | |
| 510 | marker_kwds["radius"] = marker_kwds.get("radius", 2) | |
| 511 | marker_kwds["fill"] = marker_kwds.get("fill", True) | |
| 512 | marker = folium.CircleMarker(**marker_kwds) | |
| 513 | else: | |
| 514 | raise ValueError( | |
| 515 | "Only 'marker', 'circle', and 'circle_marker' are " | |
| 516 | "supported as marker values" | |
| 517 | ) | |
| 518 | ||
| 519 | # remove additional geometries | |
| 520 | if isinstance(gdf, geopandas.GeoDataFrame): | |
| 521 | non_active_geoms = [ | |
| 522 | name | |
| 523 | for name, val in (gdf.dtypes == "geometry").items() | |
| 524 | if val and name != gdf.geometry.name | |
| 525 | ] | |
| 526 | gdf = gdf.drop(columns=non_active_geoms) | |
| 527 | ||
| 528 | # preprare tooltip and popup | |
| 529 | if isinstance(gdf, geopandas.GeoDataFrame): | |
| 530 | # add named index to the tooltip | |
| 531 | if gdf.index.name is not None: | |
| 532 | gdf = gdf.reset_index() | |
| 533 | # specify fields to show in the tooltip | |
| 534 | tooltip = _tooltip_popup("tooltip", tooltip, gdf, **tooltip_kwds) | |
| 535 | popup = _tooltip_popup("popup", popup, gdf, **popup_kwds) | |
| 536 | else: | |
| 537 | tooltip = None | |
| 538 | popup = None | |
| 539 | ||
| 540 | # add dataframe to map | |
| 541 | folium.GeoJson( | |
| 542 | gdf.__geo_interface__, | |
| 543 | tooltip=tooltip, | |
| 544 | popup=popup, | |
| 545 | marker=marker, | |
| 546 | style_function=style_function, | |
| 547 | highlight_function=highlight_function, | |
| 548 | **kwargs, | |
| 549 | ).add_to(m) | |
| 550 | ||
| 551 | if legend: | |
| 552 | # NOTE: overlaps will be resolved in branca #88 | |
| 553 | caption = column if not column == "__plottable_column" else "" | |
| 554 | caption = legend_kwds.pop("caption", caption) | |
| 555 | if categorical: | |
| 556 | categories = cat.categories.to_list() | |
| 557 | legend_colors = legend_colors.tolist() | |
| 558 | ||
| 559 | if nan_idx.any() and nan_color: | |
| 560 | categories.append(missing_kwds.pop("label", "NaN")) | |
| 561 | legend_colors.append(nan_color) | |
| 562 | ||
| 563 | _categorical_legend(m, caption, categories, legend_colors) | |
| 564 | elif column is not None: | |
| 565 | ||
| 566 | cbar = legend_kwds.pop("colorbar", True) | |
| 567 | colormap_kwds = {} | |
| 568 | if "max_labels" in legend_kwds: | |
| 569 | colormap_kwds["max_labels"] = legend_kwds.pop("max_labels") | |
| 570 | if scheme: | |
| 571 | cb_colors = np.apply_along_axis( | |
| 572 | colors.to_hex, 1, cm.get_cmap(cmap, binning.k)(range(binning.k)) | |
| 573 | ) | |
| 574 | if cbar: | |
| 575 | if legend_kwds.pop("scale", True): | |
| 576 | index = [vmin] + binning.bins.tolist() | |
| 577 | else: | |
| 578 | index = None | |
| 579 | colorbar = bc.colormap.StepColormap( | |
| 580 | cb_colors, | |
| 581 | vmin=vmin, | |
| 582 | vmax=vmax, | |
| 583 | caption=caption, | |
| 584 | index=index, | |
| 585 | **colormap_kwds, | |
| 586 | ) | |
| 587 | else: | |
| 588 | fmt = legend_kwds.pop("fmt", "{:.2f}") | |
| 589 | if "labels" in legend_kwds: | |
| 590 | categories = legend_kwds["labels"] | |
| 591 | else: | |
| 592 | categories = binning.get_legend_classes(fmt) | |
| 593 | show_interval = legend_kwds.pop("interval", False) | |
| 594 | if not show_interval: | |
| 595 | categories = [c[1:-1] for c in categories] | |
| 596 | ||
| 597 | if nan_idx.any() and nan_color: | |
| 598 | categories.append(missing_kwds.pop("label", "NaN")) | |
| 599 | cb_colors = np.append(cb_colors, nan_color) | |
| 600 | _categorical_legend(m, caption, categories, cb_colors) | |
| 601 | ||
| 602 | else: | |
| 603 | if isinstance(cmap, bc.colormap.ColorMap): | |
| 604 | colorbar = cmap | |
| 605 | else: | |
| 606 | ||
| 607 | mp_cmap = cm.get_cmap(cmap) | |
| 608 | cb_colors = np.apply_along_axis( | |
| 609 | colors.to_hex, 1, mp_cmap(range(mp_cmap.N)) | |
| 610 | ) | |
| 611 | # linear legend | |
| 612 | if mp_cmap.N > 20: | |
| 613 | colorbar = bc.colormap.LinearColormap( | |
| 614 | cb_colors, | |
| 615 | vmin=vmin, | |
| 616 | vmax=vmax, | |
| 617 | caption=caption, | |
| 618 | **colormap_kwds, | |
| 619 | ) | |
| 620 | ||
| 621 | # steps | |
| 622 | else: | |
| 623 | colorbar = bc.colormap.StepColormap( | |
| 624 | cb_colors, | |
| 625 | vmin=vmin, | |
| 626 | vmax=vmax, | |
| 627 | caption=caption, | |
| 628 | **colormap_kwds, | |
| 629 | ) | |
| 630 | ||
| 631 | if cbar: | |
| 632 | if nan_idx.any() and nan_color: | |
| 633 | _categorical_legend( | |
| 634 | m, "", [missing_kwds.pop("label", "NaN")], [nan_color] | |
| 635 | ) | |
| 636 | m.add_child(colorbar) | |
| 637 | ||
| 638 | return m | |
| 639 | ||
| 640 | ||
| 641 | def _tooltip_popup(type, fields, gdf, **kwds): | |
| 642 | """get tooltip or popup""" | |
| 643 | import folium | |
| 644 | ||
| 645 | # specify fields to show in the tooltip | |
| 646 | if fields is False or fields is None or fields == 0: | |
| 647 | return None | |
| 648 | else: | |
| 649 | if fields is True: | |
| 650 | fields = gdf.columns.drop(gdf.geometry.name).to_list() | |
| 651 | elif isinstance(fields, int): | |
| 652 | fields = gdf.columns.drop(gdf.geometry.name).to_list()[:fields] | |
| 653 | elif isinstance(fields, str): | |
| 654 | fields = [fields] | |
| 655 | ||
| 656 | for field in ["__plottable_column", "__folium_color"]: | |
| 657 | if field in fields: | |
| 658 | fields.remove(field) | |
| 659 | ||
| 660 | # Cast fields to str | |
| 661 | fields = list(map(str, fields)) | |
| 662 | if type == "tooltip": | |
| 663 | return folium.GeoJsonTooltip(fields, **kwds) | |
| 664 | elif type == "popup": | |
| 665 | return folium.GeoJsonPopup(fields, **kwds) | |
| 666 | ||
| 667 | ||
| 668 | def _categorical_legend(m, title, categories, colors): | |
| 669 | """ | |
| 670 | Add categorical legend to a map | |
| 671 | ||
| 672 | The implementation is using the code originally written by Michel Metran | |
| 673 | (@michelmetran) and released on GitHub | |
| 674 | (https://github.com/michelmetran/package_folium) under MIT license. | |
| 675 | ||
| 676 | Copyright (c) 2020 Michel Metran | |
| 677 | ||
| 678 | Parameters | |
| 679 | ---------- | |
| 680 | m : folium.Map | |
| 681 | Existing map instance on which to draw the plot | |
| 682 | title : str | |
| 683 | title of the legend (e.g. column name) | |
| 684 | categories : list-like | |
| 685 | list of categories | |
| 686 | colors : list-like | |
| 687 | list of colors (in the same order as categories) | |
| 688 | """ | |
| 689 | ||
| 690 | # Header to Add | |
| 691 | head = """ | |
| 692 | {% macro header(this, kwargs) %} | |
| 693 | <script src="https://code.jquery.com/ui/1.12.1/jquery-ui.js"></script> | |
| 694 | <script>$( function() { | |
| 695 | $( ".maplegend" ).draggable({ | |
| 696 | start: function (event, ui) { | |
| 697 | $(this).css({ | |
| 698 | right: "auto", | |
| 699 | top: "auto", | |
| 700 | bottom: "auto" | |
| 701 | }); | |
| 702 | } | |
| 703 | }); | |
| 704 | }); | |
| 705 | </script> | |
| 706 | <style type='text/css'> | |
| 707 | .maplegend { | |
| 708 | position: absolute; | |
| 709 | z-index:9999; | |
| 710 | background-color: rgba(255, 255, 255, .8); | |
| 711 | border-radius: 5px; | |
| 712 | box-shadow: 0 0 15px rgba(0,0,0,0.2); | |
| 713 | padding: 10px; | |
| 714 | font: 12px/14px Arial, Helvetica, sans-serif; | |
| 715 | right: 10px; | |
| 716 | bottom: 20px; | |
| 717 | } | |
| 718 | .maplegend .legend-title { | |
| 719 | text-align: left; | |
| 720 | margin-bottom: 5px; | |
| 721 | font-weight: bold; | |
| 722 | } | |
| 723 | .maplegend .legend-scale ul { | |
| 724 | margin: 0; | |
| 725 | margin-bottom: 0px; | |
| 726 | padding: 0; | |
| 727 | float: left; | |
| 728 | list-style: none; | |
| 729 | } | |
| 730 | .maplegend .legend-scale ul li { | |
| 731 | list-style: none; | |
| 732 | margin-left: 0; | |
| 733 | line-height: 16px; | |
| 734 | margin-bottom: 2px; | |
| 735 | } | |
| 736 | .maplegend ul.legend-labels li span { | |
| 737 | display: block; | |
| 738 | float: left; | |
| 739 | height: 14px; | |
| 740 | width: 14px; | |
| 741 | margin-right: 5px; | |
| 742 | margin-left: 0; | |
| 743 | border: 0px solid #ccc; | |
| 744 | } | |
| 745 | .maplegend .legend-source { | |
| 746 | color: #777; | |
| 747 | clear: both; | |
| 748 | } | |
| 749 | .maplegend a { | |
| 750 | color: #777; | |
| 751 | } | |
| 752 | </style> | |
| 753 | {% endmacro %} | |
| 754 | """ | |
| 755 | import branca as bc | |
| 756 | ||
| 757 | # Add CSS (on Header) | |
| 758 | macro = bc.element.MacroElement() | |
| 759 | macro._template = bc.element.Template(head) | |
| 760 | m.get_root().add_child(macro) | |
| 761 | ||
| 762 | body = f""" | |
| 763 | <div id='maplegend {title}' class='maplegend'> | |
| 764 | <div class='legend-title'>{title}</div> | |
| 765 | <div class='legend-scale'> | |
| 766 | <ul class='legend-labels'>""" | |
| 767 | ||
| 768 | # Loop Categories | |
| 769 | for label, color in zip(categories, colors): | |
| 770 | body += f""" | |
| 771 | <li><span style='background:{color}'></span>{label}</li>""" | |
| 772 | ||
| 773 | body += """ | |
| 774 | </ul> | |
| 775 | </div> | |
| 776 | </div> | |
| 777 | """ | |
| 778 | ||
| 779 | # Add Body | |
| 780 | body = bc.element.Element(body, "legend") | |
| 781 | m.get_root().html.add_child(body) | |
| 782 | ||
| 783 | ||
| 784 | def _explore_geoseries( | |
| 785 | s, | |
| 786 | color=None, | |
| 787 | m=None, | |
| 788 | tiles="OpenStreetMap", | |
| 789 | attr=None, | |
| 790 | highlight=True, | |
| 791 | width="100%", | |
| 792 | height="100%", | |
| 793 | control_scale=True, | |
| 794 | marker_type=None, | |
| 795 | marker_kwds={}, | |
| 796 | style_kwds={}, | |
| 797 | highlight_kwds={}, | |
| 798 | **kwargs, | |
| 799 | ): | |
| 800 | """Interactive map based on GeoPandas and folium/leaflet.js | |
| 801 | ||
| 802 | Generate an interactive leaflet map based on :class:`~geopandas.GeoSeries` | |
| 803 | ||
| 804 | Parameters | |
| 805 | ---------- | |
| 806 | color : str, array-like (default None) | |
| 807 | Named color or a list-like of colors (named or hex). | |
| 808 | m : folium.Map (default None) | |
| 809 | Existing map instance on which to draw the plot. | |
| 810 | tiles : str, xyzservices.TileProvider (default 'OpenStreetMap Mapnik') | |
| 811 | Map tileset to use. Can choose from the list supported by folium, query a | |
| 812 | :class:`xyzservices.TileProvider` by a name from ``xyzservices.providers``, | |
| 813 | pass :class:`xyzservices.TileProvider` object or pass custom XYZ URL. | |
| 814 | The current list of built-in providers (when ``xyzservices`` is not available): | |
| 815 | ||
| 816 | ``["OpenStreetMap", "Stamen Terrain", “Stamen Toner", “Stamen Watercolor" | |
| 817 | "CartoDB positron", “CartoDB dark_matter"]`` | |
| 818 | ||
| 819 | You can pass a custom tileset to Folium by passing a Leaflet-style URL | |
| 820 | to the tiles parameter: ``http://{s}.yourtiles.com/{z}/{x}/{y}.png``. | |
| 821 | Be sure to check their terms and conditions and to provide attribution with | |
| 822 | the ``attr`` keyword. | |
| 823 | attr : str (default None) | |
| 824 | Map tile attribution; only required if passing custom tile URL. | |
| 825 | highlight : bool (default True) | |
| 826 | Enable highlight functionality when hovering over a geometry. | |
| 827 | width : pixel int or percentage string (default: '100%') | |
| 828 | Width of the folium :class:`~folium.folium.Map`. If the argument | |
| 829 | m is given explicitly, width is ignored. | |
| 830 | height : pixel int or percentage string (default: '100%') | |
| 831 | Height of the folium :class:`~folium.folium.Map`. If the argument | |
| 832 | m is given explicitly, height is ignored. | |
| 833 | control_scale : bool, (default True) | |
| 834 | Whether to add a control scale on the map. | |
| 835 | marker_type : str, folium.Circle, folium.CircleMarker, folium.Marker (default None) | |
| 836 | Allowed string options are ('marker', 'circle', 'circle_marker'). Defaults to | |
| 837 | folium.Marker. | |
| 838 | marker_kwds: dict (default {}) | |
| 839 | Additional keywords to be passed to the selected ``marker_type``, e.g.: | |
| 840 | ||
| 841 | radius : float | |
| 842 | Radius of the circle, in meters (for ``'circle'``) or pixels | |
| 843 | (for ``circle_marker``). | |
| 844 | icon : folium.map.Icon | |
| 845 | the :class:`folium.map.Icon` object to use to render the marker. | |
| 846 | draggable : bool (default False) | |
| 847 | Set to True to be able to drag the marker around the map. | |
| 848 | ||
| 849 | style_kwds : dict (default {}) | |
| 850 | Additional style to be passed to folium ``style_function``: | |
| 851 | ||
| 852 | stroke : bool (default True) | |
| 853 | Whether to draw stroke along the path. Set it to ``False`` to | |
| 854 | disable borders on polygons or circles. | |
| 855 | color : str | |
| 856 | Stroke color | |
| 857 | weight : int | |
| 858 | Stroke width in pixels | |
| 859 | opacity : float (default 1.0) | |
| 860 | Stroke opacity | |
| 861 | fill : boolean (default True) | |
| 862 | Whether to fill the path with color. Set it to ``False`` to | |
| 863 | disable filling on polygons or circles. | |
| 864 | fillColor : str | |
| 865 | Fill color. Defaults to the value of the color option | |
| 866 | fillOpacity : float (default 0.5) | |
| 867 | Fill opacity. | |
| 868 | ||
| 869 | Plus all supported by :func:`folium.vector_layers.path_options`. See the | |
| 870 | documentation of :class:`folium.features.GeoJson` for details. | |
| 871 | ||
| 872 | highlight_kwds : dict (default {}) | |
| 873 | Style to be passed to folium highlight_function. Uses the same keywords | |
| 874 | as ``style_kwds``. When empty, defaults to ``{"fillOpacity": 0.75}``. | |
| 875 | ||
| 876 | **kwargs : dict | |
| 877 | Additional options to be passed on to the folium. | |
| 878 | ||
| 879 | Returns | |
| 880 | ------- | |
| 881 | m : folium.folium.Map | |
| 882 | folium :class:`~folium.folium.Map` instance | |
| 883 | ||
| 884 | """ | |
| 885 | return _explore( | |
| 886 | s, | |
| 887 | color=color, | |
| 888 | m=m, | |
| 889 | tiles=tiles, | |
| 890 | attr=attr, | |
| 891 | highlight=highlight, | |
| 892 | width=width, | |
| 893 | height=height, | |
| 894 | control_scale=control_scale, | |
| 895 | marker_type=marker_type, | |
| 896 | marker_kwds=marker_kwds, | |
| 897 | style_kwds=style_kwds, | |
| 898 | highlight_kwds=highlight_kwds, | |
| 899 | **kwargs, | |
| 900 | ) |
| 3 | 3 | import numpy as np |
| 4 | 4 | import pandas as pd |
| 5 | 5 | from pandas import DataFrame, Series |
| 6 | from pandas.core.accessor import CachedAccessor | |
| 6 | 7 | |
| 7 | 8 | from shapely.geometry import mapping, shape |
| 8 | 9 | from shapely.geometry.base import BaseGeometry |
| 9 | 10 | |
| 10 | ||
| 11 | 11 | from pyproj import CRS |
| 12 | 12 | |
| 13 | 13 | from geopandas.array import GeometryArray, GeometryDtype, from_shapely, to_wkb, to_wkt |
| 14 | 14 | from geopandas.base import GeoPandasBase, is_geometry_type |
| 15 | from geopandas.geoseries import GeoSeries, inherit_doc | |
| 15 | from geopandas.geoseries import GeoSeries | |
| 16 | 16 | import geopandas.io |
| 17 | from geopandas.plotting import plot_dataframe | |
| 17 | from geopandas.explore import _explore | |
| 18 | 18 | from . import _compat as compat |
| 19 | from ._decorator import doc | |
| 19 | 20 | |
| 20 | 21 | |
| 21 | 22 | DEFAULT_GEO_COLUMN_NAME = "geometry" |
| 32 | 33 | """ |
| 33 | 34 | if is_geometry_type(data): |
| 34 | 35 | if isinstance(data, Series): |
| 35 | return GeoSeries(data) | |
| 36 | data = GeoSeries(data) | |
| 37 | if data.crs is None: | |
| 38 | data.crs = crs | |
| 36 | 39 | return data |
| 37 | 40 | else: |
| 38 | 41 | if isinstance(data, Series): |
| 41 | 44 | else: |
| 42 | 45 | out = from_shapely(data, crs=crs) |
| 43 | 46 | return out |
| 47 | ||
| 48 | ||
| 49 | def _crs_mismatch_warning(): | |
| 50 | # TODO: raise error in 0.9 or 0.10. | |
| 51 | warnings.warn( | |
| 52 | "CRS mismatch between CRS of the passed geometries " | |
| 53 | "and 'crs'. Use 'GeoDataFrame.set_crs(crs, " | |
| 54 | "allow_override=True)' to overwrite CRS or " | |
| 55 | "'GeoDataFrame.to_crs(crs)' to reproject geometries. " | |
| 56 | "CRS mismatch will raise an error in the future versions " | |
| 57 | "of GeoPandas.", | |
| 58 | FutureWarning, | |
| 59 | stacklevel=3, | |
| 60 | ) | |
| 44 | 61 | |
| 45 | 62 | |
| 46 | 63 | class GeoDataFrame(GeoPandasBase, DataFrame): |
| 99 | 116 | |
| 100 | 117 | _geometry_column_name = DEFAULT_GEO_COLUMN_NAME |
| 101 | 118 | |
| 102 | def __init__(self, *args, geometry=None, crs=None, **kwargs): | |
| 119 | def __init__(self, data=None, *args, geometry=None, crs=None, **kwargs): | |
| 103 | 120 | with compat.ignore_shapely2_warnings(): |
| 104 | super(GeoDataFrame, self).__init__(*args, **kwargs) | |
| 121 | super().__init__(data, *args, **kwargs) | |
| 105 | 122 | |
| 106 | 123 | # need to set this before calling self['geometry'], because |
| 107 | 124 | # getitem accesses crs |
| 113 | 130 | # but within a try/except because currently non-geometries are |
| 114 | 131 | # allowed in that case |
| 115 | 132 | # TODO do we want to raise / return normal DataFrame in this case? |
| 133 | ||
| 134 | # if gdf passed in and geo_col is set, we use that for geometry | |
| 135 | if geometry is None and isinstance(data, GeoDataFrame): | |
| 136 | self._geometry_column_name = data._geometry_column_name | |
| 137 | if crs is not None and data.crs != crs: | |
| 138 | _crs_mismatch_warning() | |
| 139 | # TODO: raise error in 0.9 or 0.10. | |
| 140 | return | |
| 141 | ||
| 116 | 142 | if geometry is None and "geometry" in self.columns: |
| 143 | # Check for multiple columns with name "geometry". If there are, | |
| 144 | # self["geometry"] is a gdf and constructor gets recursively recalled | |
| 145 | # by pandas internals trying to access this | |
| 146 | if (self.columns == "geometry").sum() > 1: | |
| 147 | raise ValueError( | |
| 148 | "GeoDataFrame does not support multiple columns " | |
| 149 | "using the geometry column name 'geometry'." | |
| 150 | ) | |
| 151 | ||
| 117 | 152 | # only if we have actual geometry values -> call set_geometry |
| 118 | 153 | index = self.index |
| 119 | 154 | try: |
| 123 | 158 | and crs |
| 124 | 159 | and not self["geometry"].values.crs == crs |
| 125 | 160 | ): |
| 126 | warnings.warn( | |
| 127 | "CRS mismatch between CRS of the passed geometries " | |
| 128 | "and 'crs'. Use 'GeoDataFrame.set_crs(crs, " | |
| 129 | "allow_override=True)' to overwrite CRS or " | |
| 130 | "'GeoDataFrame.to_crs(crs)' to reproject geometries. " | |
| 131 | "CRS mismatch will raise an error in the future versions " | |
| 132 | "of GeoPandas.", | |
| 133 | FutureWarning, | |
| 134 | stacklevel=2, | |
| 135 | ) | |
| 161 | _crs_mismatch_warning() | |
| 136 | 162 | # TODO: raise error in 0.9 or 0.10. |
| 137 | 163 | self["geometry"] = _ensure_geometry(self["geometry"].values, crs) |
| 138 | 164 | except TypeError: |
| 152 | 178 | and crs |
| 153 | 179 | and not geometry.crs == crs |
| 154 | 180 | ): |
| 155 | warnings.warn( | |
| 156 | "CRS mismatch between CRS of the passed geometries " | |
| 157 | "and 'crs'. Use 'GeoDataFrame.set_crs(crs, " | |
| 158 | "allow_override=True)' to overwrite CRS or " | |
| 159 | "'GeoDataFrame.to_crs(crs)' to reproject geometries. " | |
| 160 | "CRS mismatch will raise an error in the future versions " | |
| 161 | "of GeoPandas.", | |
| 162 | FutureWarning, | |
| 163 | stacklevel=2, | |
| 164 | ) | |
| 181 | _crs_mismatch_warning() | |
| 165 | 182 | # TODO: raise error in 0.9 or 0.10. |
| 166 | 183 | self.set_geometry(geometry, inplace=True) |
| 167 | 184 | |
| 178 | 195 | if attr == "geometry": |
| 179 | 196 | object.__setattr__(self, attr, val) |
| 180 | 197 | else: |
| 181 | super(GeoDataFrame, self).__setattr__(attr, val) | |
| 198 | super().__setattr__(attr, val) | |
| 182 | 199 | |
| 183 | 200 | def _get_geometry(self): |
| 184 | 201 | if self._geometry_column_name not in self: |
| 267 | 284 | raise ValueError("Must pass array with one dimension only.") |
| 268 | 285 | else: |
| 269 | 286 | try: |
| 270 | level = frame[col].values | |
| 287 | level = frame[col] | |
| 271 | 288 | except KeyError: |
| 272 | 289 | raise ValueError("Unknown column %s" % col) |
| 273 | 290 | except Exception: |
| 274 | 291 | raise |
| 292 | if isinstance(level, DataFrame): | |
| 293 | raise ValueError( | |
| 294 | "GeoDataFrame does not support setting the geometry column where " | |
| 295 | "the column name is shared by multiple columns." | |
| 296 | ) | |
| 297 | ||
| 275 | 298 | if drop: |
| 276 | 299 | to_remove = col |
| 277 | 300 | geo_column_name = self._geometry_column_name |
| 428 | 451 | def from_dict(cls, data, geometry=None, crs=None, **kwargs): |
| 429 | 452 | """ |
| 430 | 453 | Construct GeoDataFrame from dict of array-like or dicts by |
| 431 | overiding DataFrame.from_dict method with geometry and crs | |
| 454 | overriding DataFrame.from_dict method with geometry and crs | |
| 432 | 455 | |
| 433 | 456 | Parameters |
| 434 | 457 | ---------- |
| 635 | 658 | PostGIS |
| 636 | 659 | |
| 637 | 660 | >>> from sqlalchemy import create_engine # doctest: +SKIP |
| 638 | >>> db_connection_url = "postgres://myusername:mypassword@myhost:5432/mydb" | |
| 661 | >>> db_connection_url = "postgresql://myusername:mypassword@myhost:5432/mydb" | |
| 639 | 662 | >>> con = create_engine(db_connection_url) # doctest: +SKIP |
| 640 | 663 | >>> sql = "SELECT geom, highway FROM roads" |
| 641 | 664 | >>> df = geopandas.GeoDataFrame.from_postgis(sql, con) # doctest: +SKIP |
| 769 | 792 | na : str, optional |
| 770 | 793 | Options are {'null', 'drop', 'keep'}, default 'null'. |
| 771 | 794 | Indicates how to output missing (NaN) values in the GeoDataFrame |
| 772 | * null: ouput the missing entries as JSON null | |
| 773 | * drop: remove the property from the feature. This applies to | |
| 774 | each feature individually so that features may have | |
| 775 | different properties | |
| 776 | * keep: output the missing entries as NaN | |
| 795 | ||
| 796 | - null: output the missing entries as JSON null | |
| 797 | - drop: remove the property from the feature. This applies to each feature \ | |
| 798 | individually so that features may have different properties | |
| 799 | - keep: output the missing entries as NaN | |
| 800 | ||
| 777 | 801 | show_bbox : bool, optional |
| 778 | 802 | Include bbox (bounds) in the geojson. Default False. |
| 779 | 803 | drop_id : bool, default: False |
| 808 | 832 | |
| 809 | 833 | ids = np.array(self.index, copy=False) |
| 810 | 834 | geometries = np.array(self[self._geometry_column_name], copy=False) |
| 835 | ||
| 836 | if not self.columns.is_unique: | |
| 837 | raise ValueError("GeoDataFrame cannot contain duplicated column names.") | |
| 811 | 838 | |
| 812 | 839 | properties_cols = self.columns.difference([self._geometry_column_name]) |
| 813 | 840 | |
| 955 | 982 | compression : {'snappy', 'gzip', 'brotli', None}, default 'snappy' |
| 956 | 983 | Name of the compression to use. Use ``None`` for no compression. |
| 957 | 984 | kwargs |
| 958 | Additional keyword arguments passed to to pyarrow.parquet.write_table(). | |
| 985 | Additional keyword arguments passed to :func:`pyarrow.parquet.write_table`. | |
| 959 | 986 | |
| 960 | 987 | Examples |
| 961 | 988 | -------- |
| 1003 | 1030 | Name of the compression to use. Use ``"uncompressed"`` for no |
| 1004 | 1031 | compression. By default uses LZ4 if available, otherwise uncompressed. |
| 1005 | 1032 | kwargs |
| 1006 | Additional keyword arguments passed to to pyarrow.feather.write_feather(). | |
| 1033 | Additional keyword arguments passed to to | |
| 1034 | :func:`pyarrow.feather.write_feather`. | |
| 1007 | 1035 | |
| 1008 | 1036 | Examples |
| 1009 | 1037 | -------- |
| 1020 | 1048 | |
| 1021 | 1049 | _to_feather(self, path, index=index, compression=compression, **kwargs) |
| 1022 | 1050 | |
| 1023 | def to_file( | |
| 1024 | self, filename, driver="ESRI Shapefile", schema=None, index=None, **kwargs | |
| 1025 | ): | |
| 1051 | def to_file(self, filename, driver=None, schema=None, index=None, **kwargs): | |
| 1026 | 1052 | """Write the ``GeoDataFrame`` to a file. |
| 1027 | 1053 | |
| 1028 | 1054 | By default, an ESRI shapefile is written, but any OGR data source |
| 1036 | 1062 | ---------- |
| 1037 | 1063 | filename : string |
| 1038 | 1064 | File path or file handle to write to. |
| 1039 | driver : string, default: 'ESRI Shapefile' | |
| 1065 | driver : string, default None | |
| 1040 | 1066 | The OGR format driver used to write the vector file. |
| 1067 | If not specified, it attempts to infer it from the file extension. | |
| 1068 | If no extension is specified, it saves ESRI Shapefile to a folder. | |
| 1041 | 1069 | schema : dict, default: None |
| 1042 | 1070 | If specified, the schema dictionary is passed to Fiona to |
| 1043 | 1071 | better control how the file is written. |
| 1295 | 1323 | GeoSeries. If it's a DataFrame with a 'geometry' column, return a |
| 1296 | 1324 | GeoDataFrame. |
| 1297 | 1325 | """ |
| 1298 | result = super(GeoDataFrame, self).__getitem__(key) | |
| 1326 | result = super().__getitem__(key) | |
| 1299 | 1327 | geo_col = self._geometry_column_name |
| 1300 | 1328 | if isinstance(result, Series) and isinstance(result.dtype, GeometryDtype): |
| 1301 | 1329 | result.__class__ = GeoSeries |
| 1316 | 1344 | value = [value] * self.shape[0] |
| 1317 | 1345 | try: |
| 1318 | 1346 | value = _ensure_geometry(value, crs=self.crs) |
| 1347 | self._crs = value.crs | |
| 1319 | 1348 | except TypeError: |
| 1320 | 1349 | warnings.warn("Geometry column does not contain geometry.") |
| 1321 | super(GeoDataFrame, self).__setitem__(key, value) | |
| 1350 | super().__setitem__(key, value) | |
| 1322 | 1351 | |
| 1323 | 1352 | # |
| 1324 | 1353 | # Implement pandas methods |
| 1355 | 1384 | result.__class__ = DataFrame |
| 1356 | 1385 | return result |
| 1357 | 1386 | |
| 1358 | @inherit_doc(pd.DataFrame) | |
| 1387 | @doc(pd.DataFrame) | |
| 1359 | 1388 | def apply(self, func, axis=0, raw=False, result_type=None, args=(), **kwargs): |
| 1360 | 1389 | result = super().apply( |
| 1361 | 1390 | func, axis=axis, raw=raw, result_type=result_type, args=args, **kwargs |
| 1363 | 1392 | if ( |
| 1364 | 1393 | isinstance(result, GeoDataFrame) |
| 1365 | 1394 | and self._geometry_column_name in result.columns |
| 1366 | and any(isinstance(t, GeometryDtype) for t in result.dtypes) | |
| 1395 | and isinstance(result[self._geometry_column_name].dtype, GeometryDtype) | |
| 1367 | 1396 | ): |
| 1397 | # apply calls _constructor which resets geom col name to geometry | |
| 1398 | result._geometry_column_name = self._geometry_column_name | |
| 1368 | 1399 | if self.crs is not None and result.crs is None: |
| 1369 | 1400 | result.set_crs(self.crs, inplace=True) |
| 1370 | 1401 | return result |
| 1374 | 1405 | return GeoDataFrame |
| 1375 | 1406 | |
| 1376 | 1407 | def __finalize__(self, other, method=None, **kwargs): |
| 1377 | """propagate metadata from other to self """ | |
| 1408 | """propagate metadata from other to self""" | |
| 1378 | 1409 | self = super().__finalize__(other, method=method, **kwargs) |
| 1379 | 1410 | |
| 1380 | 1411 | # merge operation: using metadata of the left object |
| 1386 | 1417 | for name in self._metadata: |
| 1387 | 1418 | object.__setattr__(self, name, getattr(other.objs[0], name, None)) |
| 1388 | 1419 | |
| 1420 | if (self.columns == self._geometry_column_name).sum() > 1: | |
| 1421 | raise ValueError( | |
| 1422 | "Concat operation has resulted in multiple columns using " | |
| 1423 | f"the geometry column name '{self._geometry_column_name}'.\n" | |
| 1424 | f"Please ensure this column from the first DataFrame is not " | |
| 1425 | f"repeated." | |
| 1426 | ) | |
| 1389 | 1427 | return self |
| 1390 | 1428 | |
| 1391 | 1429 | def dissolve( |
| 1512 | 1550 | return aggregated |
| 1513 | 1551 | |
| 1514 | 1552 | # overrides the pandas native explode method to break up features geometrically |
| 1515 | def explode(self, column=None, **kwargs): | |
| 1553 | def explode(self, column=None, ignore_index=False, index_parts=None, **kwargs): | |
| 1516 | 1554 | """ |
| 1517 | 1555 | Explode muti-part geometries into multiple single geometries. |
| 1518 | 1556 | |
| 1520 | 1558 | multiple rows with single geometries, thereby increasing the vertical |
| 1521 | 1559 | size of the GeoDataFrame. |
| 1522 | 1560 | |
| 1523 | The index of the input geodataframe is no longer unique and is | |
| 1524 | replaced with a multi-index (original index with additional level | |
| 1525 | indicating the multiple geometries: a new zero-based index for each | |
| 1526 | single part geometry per multi-part geometry). | |
| 1561 | .. note:: ignore_index requires pandas 1.1.0 or newer. | |
| 1562 | ||
| 1563 | Parameters | |
| 1564 | ---------- | |
| 1565 | column : string, default None | |
| 1566 | Column to explode. In the case of a geometry column, multi-part | |
| 1567 | geometries are converted to single-part. | |
| 1568 | If None, the active geometry column is used. | |
| 1569 | ignore_index : bool, default False | |
| 1570 | If True, the resulting index will be labelled 0, 1, …, n - 1, | |
| 1571 | ignoring `index_parts`. | |
| 1572 | index_parts : boolean, default True | |
| 1573 | If True, the resulting index will be a multi-index (original | |
| 1574 | index with an additional level indicating the multiple | |
| 1575 | geometries: a new zero-based index for each single part geometry | |
| 1576 | per multi-part geometry). | |
| 1527 | 1577 | |
| 1528 | 1578 | Returns |
| 1529 | 1579 | ------- |
| 1548 | 1598 | 0 name1 MULTIPOINT (1.00000 2.00000, 3.00000 4.00000) |
| 1549 | 1599 | 1 name2 MULTIPOINT (2.00000 1.00000, 0.00000 0.00000) |
| 1550 | 1600 | |
| 1551 | >>> exploded = gdf.explode() | |
| 1601 | >>> exploded = gdf.explode(index_parts=True) | |
| 1552 | 1602 | >>> exploded |
| 1553 | 1603 | col1 geometry |
| 1554 | 1604 | 0 0 name1 POINT (1.00000 2.00000) |
| 1556 | 1606 | 1 0 name2 POINT (2.00000 1.00000) |
| 1557 | 1607 | 1 name2 POINT (0.00000 0.00000) |
| 1558 | 1608 | |
| 1609 | >>> exploded = gdf.explode(index_parts=False) | |
| 1610 | >>> exploded | |
| 1611 | col1 geometry | |
| 1612 | 0 name1 POINT (1.00000 2.00000) | |
| 1613 | 0 name1 POINT (3.00000 4.00000) | |
| 1614 | 1 name2 POINT (2.00000 1.00000) | |
| 1615 | 1 name2 POINT (0.00000 0.00000) | |
| 1616 | ||
| 1617 | >>> exploded = gdf.explode(ignore_index=True) | |
| 1618 | >>> exploded | |
| 1619 | col1 geometry | |
| 1620 | 0 name1 POINT (1.00000 2.00000) | |
| 1621 | 1 name1 POINT (3.00000 4.00000) | |
| 1622 | 2 name2 POINT (2.00000 1.00000) | |
| 1623 | 3 name2 POINT (0.00000 0.00000) | |
| 1624 | ||
| 1559 | 1625 | See also |
| 1560 | 1626 | -------- |
| 1561 | 1627 | GeoDataFrame.dissolve : dissolve geometries into a single observation. |
| 1567 | 1633 | column = self.geometry.name |
| 1568 | 1634 | # If the specified column is not a geometry dtype use pandas explode |
| 1569 | 1635 | if not isinstance(self[column].dtype, GeometryDtype): |
| 1570 | return super(GeoDataFrame, self).explode(column, **kwargs) | |
| 1571 | # TODO: make sure index behaviour is consistent | |
| 1636 | if compat.PANDAS_GE_11: | |
| 1637 | return super().explode(column, ignore_index=ignore_index, **kwargs) | |
| 1638 | else: | |
| 1639 | return super().explode(column, **kwargs) | |
| 1640 | ||
| 1641 | if index_parts is None: | |
| 1642 | if not ignore_index: | |
| 1643 | warnings.warn( | |
| 1644 | "Currently, index_parts defaults to True, but in the future, " | |
| 1645 | "it will default to False to be consistent with Pandas. " | |
| 1646 | "Use `index_parts=True` to keep the current behavior and " | |
| 1647 | "True/False to silence the warning.", | |
| 1648 | FutureWarning, | |
| 1649 | stacklevel=2, | |
| 1650 | ) | |
| 1651 | index_parts = True | |
| 1572 | 1652 | |
| 1573 | 1653 | df_copy = self.copy() |
| 1574 | 1654 | |
| 1575 | if "level_1" in df_copy.columns: # GH1393 | |
| 1576 | df_copy = df_copy.rename(columns={"level_1": "__level_1"}) | |
| 1577 | ||
| 1578 | exploded_geom = df_copy.geometry.explode().reset_index(level=-1) | |
| 1579 | exploded_index = exploded_geom.columns[0] | |
| 1580 | ||
| 1581 | df = pd.concat( | |
| 1582 | [df_copy.drop(df_copy._geometry_column_name, axis=1), exploded_geom], axis=1 | |
| 1655 | level_str = f"level_{df_copy.index.nlevels}" | |
| 1656 | ||
| 1657 | if level_str in df_copy.columns: # GH1393 | |
| 1658 | df_copy = df_copy.rename(columns={level_str: f"__{level_str}"}) | |
| 1659 | ||
| 1660 | if index_parts: | |
| 1661 | exploded_geom = df_copy.geometry.explode(index_parts=True) | |
| 1662 | exploded_index = exploded_geom.index | |
| 1663 | exploded_geom = exploded_geom.reset_index(level=-1, drop=True) | |
| 1664 | else: | |
| 1665 | exploded_geom = df_copy.geometry.explode(index_parts=True).reset_index( | |
| 1666 | level=-1, drop=True | |
| 1667 | ) | |
| 1668 | exploded_index = exploded_geom.index | |
| 1669 | ||
| 1670 | df = ( | |
| 1671 | df_copy.drop(df_copy._geometry_column_name, axis=1) | |
| 1672 | .join(exploded_geom) | |
| 1673 | .__finalize__(self) | |
| 1583 | 1674 | ) |
| 1584 | # reset to MultiIndex, otherwise df index is only first level of | |
| 1585 | # exploded GeoSeries index. | |
| 1586 | df.set_index(exploded_index, append=True, inplace=True) | |
| 1587 | df.index.names = list(self.index.names) + [None] | |
| 1588 | ||
| 1589 | if "__level_1" in df.columns: | |
| 1590 | df = df.rename(columns={"__level_1": "level_1"}) | |
| 1675 | ||
| 1676 | if ignore_index: | |
| 1677 | df.reset_index(inplace=True, drop=True) | |
| 1678 | elif index_parts: | |
| 1679 | # reset to MultiIndex, otherwise df index is only first level of | |
| 1680 | # exploded GeoSeries index. | |
| 1681 | df.set_index(exploded_index, inplace=True) | |
| 1682 | df.index.names = list(self.index.names) + [None] | |
| 1683 | else: | |
| 1684 | df.set_index(exploded_index, inplace=True) | |
| 1685 | df.index.names = self.index.names | |
| 1686 | ||
| 1687 | if f"__{level_str}" in df.columns: | |
| 1688 | df = df.rename(columns={f"__{level_str}": level_str}) | |
| 1591 | 1689 | |
| 1592 | 1690 | geo_df = df.set_geometry(self._geometry_column_name) |
| 1593 | 1691 | return geo_df |
| 1606 | 1704 | ------- |
| 1607 | 1705 | GeoDataFrame or DataFrame |
| 1608 | 1706 | """ |
| 1609 | df = super(GeoDataFrame, self).astype(dtype, copy=copy, errors=errors, **kwargs) | |
| 1707 | df = super().astype(dtype, copy=copy, errors=errors, **kwargs) | |
| 1610 | 1708 | |
| 1611 | 1709 | try: |
| 1612 | 1710 | geoms = df[self._geometry_column_name] |
| 1617 | 1715 | # if the geometry column is converted to non-geometries or did not exist |
| 1618 | 1716 | # do not return a GeoDataFrame |
| 1619 | 1717 | return pd.DataFrame(df) |
| 1718 | ||
| 1719 | def convert_dtypes(self, *args, **kwargs): | |
| 1720 | """ | |
| 1721 | Convert columns to best possible dtypes using dtypes supporting ``pd.NA``. | |
| 1722 | ||
| 1723 | Always returns a GeoDataFrame as no conversions are applied to the | |
| 1724 | geometry column. | |
| 1725 | ||
| 1726 | See the pandas.DataFrame.convert_dtypes docstring for more details. | |
| 1727 | ||
| 1728 | Returns | |
| 1729 | ------- | |
| 1730 | GeoDataFrame | |
| 1731 | ||
| 1732 | """ | |
| 1733 | # Overridden to fix GH1870, that return type is not preserved always | |
| 1734 | # (and where it was, geometry col was not) | |
| 1735 | ||
| 1736 | if not compat.PANDAS_GE_10: | |
| 1737 | raise NotImplementedError( | |
| 1738 | "GeoDataFrame.convert_dtypes requires pandas >= 1.0" | |
| 1739 | ) | |
| 1740 | ||
| 1741 | return GeoDataFrame( | |
| 1742 | super().convert_dtypes(*args, **kwargs), | |
| 1743 | geometry=self.geometry.name, | |
| 1744 | crs=self.crs, | |
| 1745 | ) | |
| 1620 | 1746 | |
| 1621 | 1747 | def to_postgis( |
| 1622 | 1748 | self, |
| 1629 | 1755 | chunksize=None, |
| 1630 | 1756 | dtype=None, |
| 1631 | 1757 | ): |
| 1632 | ||
| 1633 | 1758 | """ |
| 1634 | 1759 | Upload GeoDataFrame into PostGIS database. |
| 1635 | 1760 | |
| 1669 | 1794 | -------- |
| 1670 | 1795 | |
| 1671 | 1796 | >>> from sqlalchemy import create_engine |
| 1672 | >>> engine = create_engine("postgres://myusername:mypassword@myhost:5432\ | |
| 1797 | >>> engine = create_engine("postgresql://myusername:mypassword@myhost:5432\ | |
| 1673 | 1798 | /mydatabase") # doctest: +SKIP |
| 1674 | 1799 | >>> gdf.to_postgis("my_table", engine) # doctest: +SKIP |
| 1675 | 1800 | |
| 1724 | 1849 | ) |
| 1725 | 1850 | return self.geometry.difference(other) |
| 1726 | 1851 | |
| 1727 | if compat.PANDAS_GE_025: | |
| 1728 | from pandas.core.accessor import CachedAccessor | |
| 1729 | ||
| 1730 | plot = CachedAccessor("plot", geopandas.plotting.GeoplotAccessor) | |
| 1731 | else: | |
| 1732 | ||
| 1733 | def plot(self, *args, **kwargs): | |
| 1734 | """Generate a plot of the geometries in the ``GeoDataFrame``. | |
| 1735 | If the ``column`` parameter is given, colors plot according to values | |
| 1736 | in that column, otherwise calls ``GeoSeries.plot()`` on the | |
| 1737 | ``geometry`` column. | |
| 1738 | Wraps the ``plot_dataframe()`` function, and documentation is copied | |
| 1739 | from there. | |
| 1740 | """ | |
| 1741 | return plot_dataframe(self, *args, **kwargs) | |
| 1742 | ||
| 1743 | plot.__doc__ = plot_dataframe.__doc__ | |
| 1852 | plot = CachedAccessor("plot", geopandas.plotting.GeoplotAccessor) | |
| 1853 | ||
| 1854 | @doc(_explore) | |
| 1855 | def explore(self, *args, **kwargs): | |
| 1856 | """Interactive map based on folium/leaflet.js""" | |
| 1857 | return _explore(self, *args, **kwargs) | |
| 1858 | ||
| 1859 | def sjoin(self, df, *args, **kwargs): | |
| 1860 | """Spatial join of two GeoDataFrames. | |
| 1861 | ||
| 1862 | See the User Guide page :doc:`../../user_guide/mergingdata` for details. | |
| 1863 | ||
| 1864 | Parameters | |
| 1865 | ---------- | |
| 1866 | df : GeoDataFrame | |
| 1867 | how : string, default 'inner' | |
| 1868 | The type of join: | |
| 1869 | ||
| 1870 | * 'left': use keys from left_df; retain only left_df geometry column | |
| 1871 | * 'right': use keys from right_df; retain only right_df geometry column | |
| 1872 | * 'inner': use intersection of keys from both dfs; retain only | |
| 1873 | left_df geometry column | |
| 1874 | ||
| 1875 | predicate : string, default 'intersects' | |
| 1876 | Binary predicate. Valid values are determined by the spatial index used. | |
| 1877 | You can check the valid values in left_df or right_df as | |
| 1878 | ``left_df.sindex.valid_query_predicates`` or | |
| 1879 | ``right_df.sindex.valid_query_predicates`` | |
| 1880 | lsuffix : string, default 'left' | |
| 1881 | Suffix to apply to overlapping column names (left GeoDataFrame). | |
| 1882 | rsuffix : string, default 'right' | |
| 1883 | Suffix to apply to overlapping column names (right GeoDataFrame). | |
| 1884 | ||
| 1885 | Examples | |
| 1886 | -------- | |
| 1887 | >>> countries = geopandas.read_file( \ | |
| 1888 | geopandas.datasets.get_path("naturalearth_lowres")) | |
| 1889 | >>> cities = geopandas.read_file( \ | |
| 1890 | geopandas.datasets.get_path("naturalearth_cities")) | |
| 1891 | >>> countries.head() # doctest: +SKIP | |
| 1892 | pop_est continent name \ | |
| 1893 | iso_a3 gdp_md_est geometry | |
| 1894 | 0 920938 Oceania Fiji FJI 8374.0 \ | |
| 1895 | MULTIPOLYGON (((180.00000 -16.06713, 180.00000... | |
| 1896 | 1 53950935 Africa Tanzania TZA 150600.0 \ | |
| 1897 | POLYGON ((33.90371 -0.95000, 34.07262 -1.05982... | |
| 1898 | 2 603253 Africa W. Sahara ESH 906.5 \ | |
| 1899 | POLYGON ((-8.66559 27.65643, -8.66512 27.58948... | |
| 1900 | 3 35623680 North America Canada CAN 1674000.0 \ | |
| 1901 | MULTIPOLYGON (((-122.84000 49.00000, -122.9742... | |
| 1902 | 4 326625791 North America United States of America USA 18560000.0 \ | |
| 1903 | MULTIPOLYGON (((-122.84000 49.00000, -120.0000... | |
| 1904 | >>> cities.head() | |
| 1905 | name geometry | |
| 1906 | 0 Vatican City POINT (12.45339 41.90328) | |
| 1907 | 1 San Marino POINT (12.44177 43.93610) | |
| 1908 | 2 Vaduz POINT (9.51667 47.13372) | |
| 1909 | 3 Luxembourg POINT (6.13000 49.61166) | |
| 1910 | 4 Palikir POINT (158.14997 6.91664) | |
| 1911 | ||
| 1912 | >>> cities_w_country_data = cities.sjoin(countries) | |
| 1913 | >>> cities_w_country_data.head() # doctest: +SKIP | |
| 1914 | name_left geometry index_right pop_est \ | |
| 1915 | continent name_right iso_a3 gdp_md_est | |
| 1916 | 0 Vatican City POINT (12.45339 41.90328) 141 62137802 \ | |
| 1917 | Europe Italy ITA 2221000.0 | |
| 1918 | 1 San Marino POINT (12.44177 43.93610) 141 62137802 \ | |
| 1919 | Europe Italy ITA 2221000.0 | |
| 1920 | 192 Rome POINT (12.48131 41.89790) 141 62137802 \ | |
| 1921 | Europe Italy ITA 2221000.0 | |
| 1922 | 2 Vaduz POINT (9.51667 47.13372) 114 8754413 \ | |
| 1923 | Europe Au stria AUT 416600.0 | |
| 1924 | 184 Vienna POINT (16.36469 48.20196) 114 8754413 \ | |
| 1925 | Europe Austria AUT 416600.0 | |
| 1926 | ||
| 1927 | Notes | |
| 1928 | ------ | |
| 1929 | Every operation in GeoPandas is planar, i.e. the potential third | |
| 1930 | dimension is not taken into account. | |
| 1931 | ||
| 1932 | See also | |
| 1933 | -------- | |
| 1934 | GeoDataFrame.sjoin_nearest : nearest neighbor join | |
| 1935 | sjoin : equivalent top-level function | |
| 1936 | """ | |
| 1937 | return geopandas.sjoin(left_df=self, right_df=df, *args, **kwargs) | |
| 1938 | ||
| 1939 | def sjoin_nearest( | |
| 1940 | self, | |
| 1941 | right, | |
| 1942 | how="inner", | |
| 1943 | max_distance=None, | |
| 1944 | lsuffix="left", | |
| 1945 | rsuffix="right", | |
| 1946 | distance_col=None, | |
| 1947 | ): | |
| 1948 | """ | |
| 1949 | Spatial join of two GeoDataFrames based on the distance between their | |
| 1950 | geometries. | |
| 1951 | ||
| 1952 | Results will include multiple output records for a single input record | |
| 1953 | where there are multiple equidistant nearest or intersected neighbors. | |
| 1954 | ||
| 1955 | See the User Guide page | |
| 1956 | https://geopandas.readthedocs.io/en/latest/docs/user_guide/mergingdata.html | |
| 1957 | for more details. | |
| 1958 | ||
| 1959 | ||
| 1960 | Parameters | |
| 1961 | ---------- | |
| 1962 | right : GeoDataFrame | |
| 1963 | how : string, default 'inner' | |
| 1964 | The type of join: | |
| 1965 | ||
| 1966 | * 'left': use keys from left_df; retain only left_df geometry column | |
| 1967 | * 'right': use keys from right_df; retain only right_df geometry column | |
| 1968 | * 'inner': use intersection of keys from both dfs; retain only | |
| 1969 | left_df geometry column | |
| 1970 | ||
| 1971 | max_distance : float, default None | |
| 1972 | Maximum distance within which to query for nearest geometry. | |
| 1973 | Must be greater than 0. | |
| 1974 | The max_distance used to search for nearest items in the tree may have a | |
| 1975 | significant impact on performance by reducing the number of input | |
| 1976 | geometries that are evaluated for nearest items in the tree. | |
| 1977 | lsuffix : string, default 'left' | |
| 1978 | Suffix to apply to overlapping column names (left GeoDataFrame). | |
| 1979 | rsuffix : string, default 'right' | |
| 1980 | Suffix to apply to overlapping column names (right GeoDataFrame). | |
| 1981 | distance_col : string, default None | |
| 1982 | If set, save the distances computed between matching geometries under a | |
| 1983 | column of this name in the joined GeoDataFrame. | |
| 1984 | ||
| 1985 | Examples | |
| 1986 | -------- | |
| 1987 | >>> countries = geopandas.read_file(geopandas.datasets.get_\ | |
| 1988 | path("naturalearth_lowres")) | |
| 1989 | >>> cities = geopandas.read_file(geopandas.datasets.get_path("naturalearth_citi\ | |
| 1990 | es")) | |
| 1991 | >>> countries.head(2).name # doctest: +SKIP | |
| 1992 | pop_est continent name \ | |
| 1993 | iso_a3 gdp_md_est geometry | |
| 1994 | 0 920938 Oceania Fiji FJI 8374.0 MULTI\ | |
| 1995 | POLYGON (((180.00000 -16.06713, 180.00000... | |
| 1996 | 1 53950935 Africa Tanzania TZA 150600.0 POLYG\ | |
| 1997 | ON ((33.90371 -0.95000, 34.07262 -1.05982... | |
| 1998 | >>> cities.head(2).name # doctest: +SKIP | |
| 1999 | name geometry | |
| 2000 | 0 Vatican City POINT (12.45339 41.90328) | |
| 2001 | 1 San Marino POINT (12.44177 43.93610) | |
| 2002 | ||
| 2003 | >>> cities_w_country_data = cities.sjoin_nearest(countries) | |
| 2004 | >>> cities_w_country_data[['name_left', 'name_right']].head(2) # doctest: +SKIP | |
| 2005 | name_left geometry index_right pop_est continent n\ | |
| 2006 | ame_right iso_a3 gdp_md_est | |
| 2007 | 0 Vatican City POINT (12.45339 41.90328) 141 62137802 Europe \ | |
| 2008 | Italy ITA 2221000.0 | |
| 2009 | 1 San Marino POINT (12.44177 43.93610) 141 62137802 Europe \ | |
| 2010 | Italy ITA 2221000.0 | |
| 2011 | ||
| 2012 | To include the distances: | |
| 2013 | ||
| 2014 | >>> cities_w_country_data = cities.sjoin_nearest(countries, \ | |
| 2015 | distance_col="distances") | |
| 2016 | >>> cities_w_country_data[["name_left", "name_right", \ | |
| 2017 | "distances"]].head(2) # doctest: +SKIP | |
| 2018 | name_left name_right distances | |
| 2019 | 0 Vatican City Italy 0.0 | |
| 2020 | 1 San Marino Italy 0.0 | |
| 2021 | ||
| 2022 | In the following example, we get multiple cities for Italy because all results | |
| 2023 | are equidistant (in this case zero because they intersect). | |
| 2024 | In fact, we get 3 results in total: | |
| 2025 | ||
| 2026 | >>> countries_w_city_data = cities.sjoin_nearest(countries, \ | |
| 2027 | distance_col="distances", how="right") | |
| 2028 | >>> italy_results = \ | |
| 2029 | countries_w_city_data[countries_w_city_data["name_left"] == "Italy"] | |
| 2030 | >>> italy_results # doctest: +SKIP | |
| 2031 | name_x name_y | |
| 2032 | 141 Vatican City Italy | |
| 2033 | 141 San Marino Italy | |
| 2034 | 141 Rome Italy | |
| 2035 | ||
| 2036 | See also | |
| 2037 | -------- | |
| 2038 | GeoDataFrame.sjoin : binary predicate joins | |
| 2039 | sjoin_nearest : equivalent top-level function | |
| 2040 | ||
| 2041 | Notes | |
| 2042 | ----- | |
| 2043 | Since this join relies on distances, results will be innaccurate | |
| 2044 | if your geometries are in a geographic CRS. | |
| 2045 | ||
| 2046 | Every operation in GeoPandas is planar, i.e. the potential third | |
| 2047 | dimension is not taken into account. | |
| 2048 | """ | |
| 2049 | return geopandas.sjoin_nearest( | |
| 2050 | self, | |
| 2051 | right, | |
| 2052 | how=how, | |
| 2053 | max_distance=max_distance, | |
| 2054 | lsuffix=lsuffix, | |
| 2055 | rsuffix=rsuffix, | |
| 2056 | distance_col=distance_col, | |
| 2057 | ) | |
| 2058 | ||
| 2059 | def clip(self, mask, keep_geom_type=False): | |
| 2060 | """Clip points, lines, or polygon geometries to the mask extent. | |
| 2061 | ||
| 2062 | Both layers must be in the same Coordinate Reference System (CRS). | |
| 2063 | The GeoDataFrame will be clipped to the full extent of the `mask` object. | |
| 2064 | ||
| 2065 | If there are multiple polygons in mask, data from the GeoDataFrame will be | |
| 2066 | clipped to the total boundary of all polygons in mask. | |
| 2067 | ||
| 2068 | Parameters | |
| 2069 | ---------- | |
| 2070 | mask : GeoDataFrame, GeoSeries, (Multi)Polygon | |
| 2071 | Polygon vector layer used to clip `gdf`. | |
| 2072 | The mask's geometry is dissolved into one geometric feature | |
| 2073 | and intersected with `gdf`. | |
| 2074 | keep_geom_type : boolean, default False | |
| 2075 | If True, return only geometries of original type in case of intersection | |
| 2076 | resulting in multiple geometry types or GeometryCollections. | |
| 2077 | If False, return all resulting geometries (potentially mixed types). | |
| 2078 | ||
| 2079 | Returns | |
| 2080 | ------- | |
| 2081 | GeoDataFrame | |
| 2082 | Vector data (points, lines, polygons) from `gdf` clipped to | |
| 2083 | polygon boundary from mask. | |
| 2084 | ||
| 2085 | See also | |
| 2086 | -------- | |
| 2087 | clip : equivalent top-level function | |
| 2088 | ||
| 2089 | Examples | |
| 2090 | -------- | |
| 2091 | Clip points (global cities) with a polygon (the South American continent): | |
| 2092 | ||
| 2093 | >>> world = geopandas.read_file( | |
| 2094 | ... geopandas.datasets.get_path('naturalearth_lowres')) | |
| 2095 | >>> south_america = world[world['continent'] == "South America"] | |
| 2096 | >>> capitals = geopandas.read_file( | |
| 2097 | ... geopandas.datasets.get_path('naturalearth_cities')) | |
| 2098 | >>> capitals.shape | |
| 2099 | (202, 2) | |
| 2100 | ||
| 2101 | >>> sa_capitals = capitals.clip(south_america) | |
| 2102 | >>> sa_capitals.shape | |
| 2103 | (12, 2) | |
| 2104 | """ | |
| 2105 | return geopandas.clip(self, mask=mask, keep_geom_type=keep_geom_type) | |
| 2106 | ||
| 2107 | def overlay(self, right, how="intersection", keep_geom_type=None, make_valid=True): | |
| 2108 | """Perform spatial overlay between GeoDataFrames. | |
| 2109 | ||
| 2110 | Currently only supports data GeoDataFrames with uniform geometry types, | |
| 2111 | i.e. containing only (Multi)Polygons, or only (Multi)Points, or a | |
| 2112 | combination of (Multi)LineString and LinearRing shapes. | |
| 2113 | Implements several methods that are all effectively subsets of the union. | |
| 2114 | ||
| 2115 | See the User Guide page :doc:`../../user_guide/set_operations` for details. | |
| 2116 | ||
| 2117 | Parameters | |
| 2118 | ---------- | |
| 2119 | right : GeoDataFrame | |
| 2120 | how : string | |
| 2121 | Method of spatial overlay: 'intersection', 'union', | |
| 2122 | 'identity', 'symmetric_difference' or 'difference'. | |
| 2123 | keep_geom_type : bool | |
| 2124 | If True, return only geometries of the same geometry type the GeoDataFrame | |
| 2125 | has, if False, return all resulting geometries. Default is None, | |
| 2126 | which will set keep_geom_type to True but warn upon dropping | |
| 2127 | geometries. | |
| 2128 | make_valid : bool, default True | |
| 2129 | If True, any invalid input geometries are corrected with a call to | |
| 2130 | `buffer(0)`, if False, a `ValueError` is raised if any input geometries | |
| 2131 | are invalid. | |
| 2132 | ||
| 2133 | Returns | |
| 2134 | ------- | |
| 2135 | df : GeoDataFrame | |
| 2136 | GeoDataFrame with new set of polygons and attributes | |
| 2137 | resulting from the overlay | |
| 2138 | ||
| 2139 | Examples | |
| 2140 | -------- | |
| 2141 | >>> from shapely.geometry import Polygon | |
| 2142 | >>> polys1 = geopandas.GeoSeries([Polygon([(0,0), (2,0), (2,2), (0,2)]), | |
| 2143 | ... Polygon([(2,2), (4,2), (4,4), (2,4)])]) | |
| 2144 | >>> polys2 = geopandas.GeoSeries([Polygon([(1,1), (3,1), (3,3), (1,3)]), | |
| 2145 | ... Polygon([(3,3), (5,3), (5,5), (3,5)])]) | |
| 2146 | >>> df1 = geopandas.GeoDataFrame({'geometry': polys1, 'df1_data':[1,2]}) | |
| 2147 | >>> df2 = geopandas.GeoDataFrame({'geometry': polys2, 'df2_data':[1,2]}) | |
| 2148 | ||
| 2149 | >>> df1.overlay(df2, how='union') | |
| 2150 | df1_data df2_data geometry | |
| 2151 | 0 1.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 2152 | 1 2.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 2153 | 2 2.0 2.0 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 2154 | 3 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 2155 | 4 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... | |
| 2156 | 5 NaN 1.0 MULTIPOLYGON (((2.00000 2.00000, 3.00000 2.000... | |
| 2157 | 6 NaN 2.0 POLYGON ((3.00000 5.00000, 5.00000 5.00000, 5.... | |
| 2158 | ||
| 2159 | >>> df1.overlay(df2, how='intersection') | |
| 2160 | df1_data df2_data geometry | |
| 2161 | 0 1 1 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 2162 | 1 2 1 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 2163 | 2 2 2 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 2164 | ||
| 2165 | >>> df1.overlay(df2, how='symmetric_difference') | |
| 2166 | df1_data df2_data geometry | |
| 2167 | 0 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 2168 | 1 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... | |
| 2169 | 2 NaN 1.0 MULTIPOLYGON (((2.00000 2.00000, 3.00000 2.000... | |
| 2170 | 3 NaN 2.0 POLYGON ((3.00000 5.00000, 5.00000 5.00000, 5.... | |
| 2171 | ||
| 2172 | >>> df1.overlay(df2, how='difference') | |
| 2173 | geometry df1_data | |
| 2174 | 0 POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... 1 | |
| 2175 | 1 MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... 2 | |
| 2176 | ||
| 2177 | >>> df1.overlay(df2, how='identity') | |
| 2178 | df1_data df2_data geometry | |
| 2179 | 0 1.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 2180 | 1 2.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 2181 | 2 2.0 2.0 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 2182 | 3 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 2183 | 4 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... | |
| 2184 | ||
| 2185 | See also | |
| 2186 | -------- | |
| 2187 | GeoDataFrame.sjoin : spatial join | |
| 2188 | overlay : equivalent top-level function | |
| 2189 | ||
| 2190 | Notes | |
| 2191 | ------ | |
| 2192 | Every operation in GeoPandas is planar, i.e. the potential third | |
| 2193 | dimension is not taken into account. | |
| 2194 | """ | |
| 2195 | return geopandas.overlay( | |
| 2196 | self, right, how=how, keep_geom_type=keep_geom_type, make_valid=make_valid | |
| 2197 | ) | |
| 1744 | 2198 | |
| 1745 | 2199 | |
| 1746 | 2200 | def _dataframe_set_geometry(self, col, drop=False, inplace=False, crs=None): |
| 10 | 10 | |
| 11 | 11 | from geopandas.base import GeoPandasBase, _delegate_property |
| 12 | 12 | from geopandas.plotting import plot_series |
| 13 | ||
| 13 | from geopandas.explore import _explore_geoseries | |
| 14 | import geopandas | |
| 15 | ||
| 16 | from . import _compat as compat | |
| 17 | from ._decorator import doc | |
| 14 | 18 | from .array import ( |
| 15 | 19 | GeometryDtype, |
| 16 | 20 | from_shapely, |
| 17 | 21 | from_wkb, |
| 18 | 22 | from_wkt, |
| 23 | points_from_xy, | |
| 19 | 24 | to_wkb, |
| 20 | 25 | to_wkt, |
| 21 | 26 | ) |
| 22 | 27 | from .base import is_geometry_type |
| 23 | from . import _compat as compat | |
| 24 | 28 | |
| 25 | 29 | |
| 26 | 30 | _SERIES_WARNING_MSG = """\ |
| 46 | 50 | return GeoSeries(data=data, index=index, crs=crs, **kwargs) |
| 47 | 51 | except TypeError: |
| 48 | 52 | return Series(data=data, index=index, **kwargs) |
| 49 | ||
| 50 | ||
| 51 | def inherit_doc(cls): | |
| 52 | """ | |
| 53 | A decorator adding a docstring from an existing method. | |
| 54 | """ | |
| 55 | ||
| 56 | def decorator(decorated): | |
| 57 | original_method = getattr(cls, decorated.__name__, None) | |
| 58 | if original_method: | |
| 59 | doc = original_method.__doc__ or "" | |
| 60 | else: | |
| 61 | doc = "" | |
| 62 | ||
| 63 | decorated.__doc__ = doc | |
| 64 | return decorated | |
| 65 | ||
| 66 | return decorator | |
| 67 | 53 | |
| 68 | 54 | |
| 69 | 55 | class GeoSeries(GeoPandasBase, Series): |
| 131 | 117 | - Lat[north]: Geodetic latitude (degree) |
| 132 | 118 | - Lon[east]: Geodetic longitude (degree) |
| 133 | 119 | Area of Use: |
| 134 | - name: World | |
| 120 | - name: World. | |
| 135 | 121 | - bounds: (-180.0, -90.0, 180.0, 90.0) |
| 136 | Datum: World Geodetic System 1984 | |
| 122 | Datum: World Geodetic System 1984 ensemble | |
| 137 | 123 | - Ellipsoid: WGS 84 |
| 138 | 124 | - Prime Meridian: Greenwich |
| 139 | 125 | |
| 203 | 189 | kwargs.pop("dtype", None) |
| 204 | 190 | # Use Series constructor to handle input data |
| 205 | 191 | with compat.ignore_shapely2_warnings(): |
| 192 | # suppress additional warning from pandas for empty data | |
| 193 | # (will always give object dtype instead of float dtype in the future, | |
| 194 | # making the `if s.empty: s = s.astype(object)` below unnecessary) | |
| 195 | empty_msg = "The default dtype for empty Series" | |
| 196 | warnings.filterwarnings("ignore", empty_msg, DeprecationWarning) | |
| 197 | warnings.filterwarnings("ignore", empty_msg, FutureWarning) | |
| 206 | 198 | s = pd.Series(data, index=index, name=name, **kwargs) |
| 207 | 199 | # prevent trying to convert non-geometry objects |
| 208 | 200 | if s.dtype != object: |
| 209 | if s.empty or data is None: | |
| 201 | if (s.empty and s.dtype == "float64") or data is None: | |
| 202 | # pd.Series with empty data gives float64 for older pandas versions | |
| 210 | 203 | s = s.astype(object) |
| 211 | 204 | else: |
| 212 | 205 | warnings.warn(_SERIES_WARNING_MSG, FutureWarning, stacklevel=2) |
| 445 | 438 | return cls._from_wkb_or_wkb(from_wkt, data, index=index, crs=crs, **kwargs) |
| 446 | 439 | |
| 447 | 440 | @classmethod |
| 441 | def from_xy(cls, x, y, z=None, index=None, crs=None, **kwargs): | |
| 442 | """ | |
| 443 | Alternate constructor to create a :class:`~geopandas.GeoSeries` of Point | |
| 444 | geometries from lists or arrays of x, y(, z) coordinates | |
| 445 | ||
| 446 | In case of geographic coordinates, it is assumed that longitude is captured | |
| 447 | by ``x`` coordinates and latitude by ``y``. | |
| 448 | ||
| 449 | Parameters | |
| 450 | ---------- | |
| 451 | x, y, z : iterable | |
| 452 | index : array-like or Index, optional | |
| 453 | The index for the GeoSeries. If not given and all coordinate inputs | |
| 454 | are Series with an equal index, that index is used. | |
| 455 | crs : value, optional | |
| 456 | Coordinate Reference System of the geometry objects. Can be anything | |
| 457 | accepted by | |
| 458 | :meth:`pyproj.CRS.from_user_input() <pyproj.crs.CRS.from_user_input>`, | |
| 459 | such as an authority string (eg "EPSG:4326") or a WKT string. | |
| 460 | **kwargs | |
| 461 | Additional arguments passed to the Series constructor, | |
| 462 | e.g. ``name``. | |
| 463 | ||
| 464 | Returns | |
| 465 | ------- | |
| 466 | GeoSeries | |
| 467 | ||
| 468 | See Also | |
| 469 | -------- | |
| 470 | GeoSeries.from_wkt | |
| 471 | points_from_xy | |
| 472 | ||
| 473 | Examples | |
| 474 | -------- | |
| 475 | ||
| 476 | >>> x = [2.5, 5, -3.0] | |
| 477 | >>> y = [0.5, 1, 1.5] | |
| 478 | >>> s = geopandas.GeoSeries.from_xy(x, y, crs="EPSG:4326") | |
| 479 | >>> s | |
| 480 | 0 POINT (2.50000 0.50000) | |
| 481 | 1 POINT (5.00000 1.00000) | |
| 482 | 2 POINT (-3.00000 1.50000) | |
| 483 | dtype: geometry | |
| 484 | """ | |
| 485 | if index is None: | |
| 486 | if ( | |
| 487 | isinstance(x, Series) | |
| 488 | and isinstance(y, Series) | |
| 489 | and x.index.equals(y.index) | |
| 490 | and (z is None or (isinstance(z, Series) and x.index.equals(z.index))) | |
| 491 | ): # check if we can reuse index | |
| 492 | index = x.index | |
| 493 | return cls(points_from_xy(x, y, z, crs=crs), index=index, crs=crs, **kwargs) | |
| 494 | ||
| 495 | @classmethod | |
| 448 | 496 | def _from_wkb_or_wkb( |
| 449 | 497 | cls, from_wkb_or_wkt_function, data, index=None, crs=None, **kwargs |
| 450 | 498 | ): |
| 484 | 532 | |
| 485 | 533 | return GeoDataFrame({"geometry": self}).__geo_interface__ |
| 486 | 534 | |
| 487 | def to_file(self, filename, driver="ESRI Shapefile", index=None, **kwargs): | |
| 535 | def to_file(self, filename, driver=None, index=None, **kwargs): | |
| 488 | 536 | """Write the ``GeoSeries`` to a file. |
| 489 | 537 | |
| 490 | 538 | By default, an ESRI shapefile is written, but any OGR data source |
| 494 | 542 | ---------- |
| 495 | 543 | filename : string |
| 496 | 544 | File path or file handle to write to. |
| 497 | driver : string, default: 'ESRI Shapefile' | |
| 545 | driver : string, default None | |
| 498 | 546 | The OGR format driver used to write the vector file. |
| 547 | If not specified, it attempts to infer it from the file extension. | |
| 548 | If no extension is specified, it saves ESRI Shapefile to a folder. | |
| 499 | 549 | index : bool, default None |
| 500 | 550 | If True, write index into one or more columns (for MultiIndex). |
| 501 | 551 | Default None writes the index into one or more columns only if |
| 547 | 597 | |
| 548 | 598 | def _wrapped_pandas_method(self, mtd, *args, **kwargs): |
| 549 | 599 | """Wrap a generic pandas method to ensure it returns a GeoSeries""" |
| 550 | val = getattr(super(GeoSeries, self), mtd)(*args, **kwargs) | |
| 600 | val = getattr(super(), mtd)(*args, **kwargs) | |
| 551 | 601 | if type(val) == Series: |
| 552 | 602 | val.__class__ = GeoSeries |
| 553 | 603 | val.crs = self.crs |
| 556 | 606 | def __getitem__(self, key): |
| 557 | 607 | return self._wrapped_pandas_method("__getitem__", key) |
| 558 | 608 | |
| 559 | @inherit_doc(pd.Series) | |
| 609 | @doc(pd.Series) | |
| 560 | 610 | def sort_index(self, *args, **kwargs): |
| 561 | 611 | return self._wrapped_pandas_method("sort_index", *args, **kwargs) |
| 562 | 612 | |
| 563 | @inherit_doc(pd.Series) | |
| 613 | @doc(pd.Series) | |
| 564 | 614 | def take(self, *args, **kwargs): |
| 565 | 615 | return self._wrapped_pandas_method("take", *args, **kwargs) |
| 566 | 616 | |
| 567 | @inherit_doc(pd.Series) | |
| 617 | @doc(pd.Series) | |
| 568 | 618 | def select(self, *args, **kwargs): |
| 569 | 619 | return self._wrapped_pandas_method("select", *args, **kwargs) |
| 570 | 620 | |
| 571 | @inherit_doc(pd.Series) | |
| 621 | @doc(pd.Series) | |
| 572 | 622 | def apply(self, func, convert_dtype=True, args=(), **kwargs): |
| 573 | 623 | result = super().apply(func, convert_dtype=convert_dtype, args=args, **kwargs) |
| 574 | 624 | if isinstance(result, GeoSeries): |
| 577 | 627 | return result |
| 578 | 628 | |
| 579 | 629 | def __finalize__(self, other, method=None, **kwargs): |
| 580 | """ propagate metadata from other to self """ | |
| 630 | """propagate metadata from other to self""" | |
| 581 | 631 | # NOTE: backported from pandas master (upcoming v0.13) |
| 582 | 632 | for name in self._metadata: |
| 583 | 633 | object.__setattr__(self, name, getattr(other, name, None)) |
| 636 | 686 | stacklevel=2, |
| 637 | 687 | ) |
| 638 | 688 | |
| 639 | return super(GeoSeries, self).isna() | |
| 689 | return super().isna() | |
| 640 | 690 | |
| 641 | 691 | def isnull(self): |
| 642 | 692 | """Alias for `isna` method. See `isna` for more detail.""" |
| 694 | 744 | UserWarning, |
| 695 | 745 | stacklevel=2, |
| 696 | 746 | ) |
| 697 | return super(GeoSeries, self).notna() | |
| 747 | return super().notna() | |
| 698 | 748 | |
| 699 | 749 | def notnull(self): |
| 700 | 750 | """Alias for `notna` method. See `notna` for more detail.""" |
| 740 | 790 | """ |
| 741 | 791 | if value is None: |
| 742 | 792 | value = BaseGeometry() |
| 743 | return super(GeoSeries, self).fillna( | |
| 744 | value=value, method=method, inplace=inplace, **kwargs | |
| 745 | ) | |
| 793 | return super().fillna(value=value, method=method, inplace=inplace, **kwargs) | |
| 746 | 794 | |
| 747 | 795 | def __contains__(self, other): |
| 748 | 796 | """Allow tests of the form "geom in s" |
| 756 | 804 | else: |
| 757 | 805 | return False |
| 758 | 806 | |
| 807 | @doc(plot_series) | |
| 759 | 808 | def plot(self, *args, **kwargs): |
| 760 | """Generate a plot of the geometries in the ``GeoSeries``. | |
| 761 | ||
| 762 | Wraps the ``plot_series()`` function, and documentation is copied from | |
| 763 | there. | |
| 764 | """ | |
| 765 | 809 | return plot_series(self, *args, **kwargs) |
| 766 | 810 | |
| 767 | plot.__doc__ = plot_series.__doc__ | |
| 768 | ||
| 769 | def explode(self): | |
| 811 | @doc(_explore_geoseries) | |
| 812 | def explore(self, *args, **kwargs): | |
| 813 | """Interactive map based on folium/leaflet.js""" | |
| 814 | return _explore_geoseries(self, *args, **kwargs) | |
| 815 | ||
| 816 | def explode(self, ignore_index=False, index_parts=None): | |
| 770 | 817 | """ |
| 771 | 818 | Explode multi-part geometries into multiple single geometries. |
| 772 | 819 | |
| 774 | 821 | This is analogous to PostGIS's ST_Dump(). The 'path' index is the |
| 775 | 822 | second level of the returned MultiIndex |
| 776 | 823 | |
| 777 | Returns | |
| 778 | ------ | |
| 824 | Parameters | |
| 825 | ---------- | |
| 826 | ignore_index : bool, default False | |
| 827 | If True, the resulting index will be labelled 0, 1, …, n - 1, | |
| 828 | ignoring `index_parts`. | |
| 829 | index_parts : boolean, default True | |
| 830 | If True, the resulting index will be a multi-index (original | |
| 831 | index with an additional level indicating the multiple | |
| 832 | geometries: a new zero-based index for each single part geometry | |
| 833 | per multi-part geometry). | |
| 834 | ||
| 835 | Returns | |
| 836 | ------- | |
| 779 | 837 | A GeoSeries with a MultiIndex. The levels of the MultiIndex are the |
| 780 | 838 | original index and a zero-based integer index that counts the |
| 781 | 839 | number of single geometries within a multi-part geometry. |
| 791 | 849 | 1 MULTIPOINT (2.00000 2.00000, 3.00000 3.00000, ... |
| 792 | 850 | dtype: geometry |
| 793 | 851 | |
| 794 | >>> s.explode() | |
| 852 | >>> s.explode(index_parts=True) | |
| 795 | 853 | 0 0 POINT (0.00000 0.00000) |
| 796 | 854 | 1 POINT (1.00000 1.00000) |
| 797 | 855 | 1 0 POINT (2.00000 2.00000) |
| 804 | 862 | GeoDataFrame.explode |
| 805 | 863 | |
| 806 | 864 | """ |
| 865 | if index_parts is None and not ignore_index: | |
| 866 | warnings.warn( | |
| 867 | "Currently, index_parts defaults to True, but in the future, " | |
| 868 | "it will default to False to be consistent with Pandas. " | |
| 869 | "Use `index_parts=True` to keep the current behavior and True/False " | |
| 870 | "to silence the warning.", | |
| 871 | FutureWarning, | |
| 872 | stacklevel=2, | |
| 873 | ) | |
| 874 | index_parts = True | |
| 807 | 875 | |
| 808 | 876 | if compat.USE_PYGEOS and compat.PYGEOS_GE_09: |
| 809 | 877 | import pygeos # noqa |
| 828 | 896 | |
| 829 | 897 | # extract original index values based on integer index |
| 830 | 898 | outer_index = self.index.take(outer_idx) |
| 831 | ||
| 832 | index = MultiIndex.from_arrays( | |
| 833 | [outer_index, inner_index], names=self.index.names + [None] | |
| 834 | ) | |
| 899 | if ignore_index: | |
| 900 | index = range(len(geometries)) | |
| 901 | ||
| 902 | elif index_parts: | |
| 903 | nlevels = outer_index.nlevels | |
| 904 | index_arrays = [ | |
| 905 | outer_index.get_level_values(lvl) for lvl in range(nlevels) | |
| 906 | ] | |
| 907 | index_arrays.append(inner_index) | |
| 908 | ||
| 909 | index = MultiIndex.from_arrays( | |
| 910 | index_arrays, names=self.index.names + [None] | |
| 911 | ) | |
| 912 | ||
| 913 | else: | |
| 914 | index = outer_index | |
| 835 | 915 | |
| 836 | 916 | return GeoSeries(geometries, index=index, crs=self.crs).__finalize__(self) |
| 837 | 917 | |
| 848 | 928 | idxs = [(idx, 0)] |
| 849 | 929 | index.extend(idxs) |
| 850 | 930 | geometries.extend(geoms) |
| 851 | index = MultiIndex.from_tuples(index, names=self.index.names + [None]) | |
| 931 | ||
| 932 | if ignore_index: | |
| 933 | index = range(len(geometries)) | |
| 934 | ||
| 935 | elif index_parts: | |
| 936 | # if self.index is a MultiIndex then index is a list of nested tuples | |
| 937 | if isinstance(self.index, MultiIndex): | |
| 938 | index = [tuple(outer) + (inner,) for outer, inner in index] | |
| 939 | index = MultiIndex.from_tuples(index, names=self.index.names + [None]) | |
| 940 | ||
| 941 | else: | |
| 942 | index = [idx for idx, _ in index] | |
| 943 | ||
| 852 | 944 | return GeoSeries(geometries, index=index, crs=self.crs).__finalize__(self) |
| 853 | 945 | |
| 854 | 946 | # |
| 1204 | 1296 | stacklevel=2, |
| 1205 | 1297 | ) |
| 1206 | 1298 | return self.difference(other) |
| 1299 | ||
| 1300 | def clip(self, mask, keep_geom_type=False): | |
| 1301 | """Clip points, lines, or polygon geometries to the mask extent. | |
| 1302 | ||
| 1303 | Both layers must be in the same Coordinate Reference System (CRS). | |
| 1304 | The GeoSeries will be clipped to the full extent of the `mask` object. | |
| 1305 | ||
| 1306 | If there are multiple polygons in mask, data from the GeoSeries will be | |
| 1307 | clipped to the total boundary of all polygons in mask. | |
| 1308 | ||
| 1309 | Parameters | |
| 1310 | ---------- | |
| 1311 | mask : GeoDataFrame, GeoSeries, (Multi)Polygon | |
| 1312 | Polygon vector layer used to clip `gdf`. | |
| 1313 | The mask's geometry is dissolved into one geometric feature | |
| 1314 | and intersected with `gdf`. | |
| 1315 | keep_geom_type : boolean, default False | |
| 1316 | If True, return only geometries of original type in case of intersection | |
| 1317 | resulting in multiple geometry types or GeometryCollections. | |
| 1318 | If False, return all resulting geometries (potentially mixed-types). | |
| 1319 | ||
| 1320 | Returns | |
| 1321 | ------- | |
| 1322 | GeoSeries | |
| 1323 | Vector data (points, lines, polygons) from `gdf` clipped to | |
| 1324 | polygon boundary from mask. | |
| 1325 | ||
| 1326 | See also | |
| 1327 | -------- | |
| 1328 | clip : top-level function for clip | |
| 1329 | ||
| 1330 | Examples | |
| 1331 | -------- | |
| 1332 | Clip points (global cities) with a polygon (the South American continent): | |
| 1333 | ||
| 1334 | >>> world = geopandas.read_file( | |
| 1335 | ... geopandas.datasets.get_path('naturalearth_lowres')) | |
| 1336 | >>> south_america = world[world['continent'] == "South America"] | |
| 1337 | >>> capitals = geopandas.read_file( | |
| 1338 | ... geopandas.datasets.get_path('naturalearth_cities')) | |
| 1339 | >>> capitals.shape | |
| 1340 | (202, 2) | |
| 1341 | ||
| 1342 | >>> sa_capitals = capitals.geometry.clip(south_america) | |
| 1343 | >>> sa_capitals.shape | |
| 1344 | (12,) | |
| 1345 | """ | |
| 1346 | return geopandas.clip(self, mask=mask, keep_geom_type=keep_geom_type) | |
| 7 | 7 | from geopandas.array import from_wkb |
| 8 | 8 | from geopandas import GeoDataFrame |
| 9 | 9 | import geopandas |
| 10 | ||
| 10 | from .file import _expand_user | |
| 11 | 11 | |
| 12 | 12 | METADATA_VERSION = "0.1.0" |
| 13 | 13 | # reference: https://github.com/geopandas/geo-arrow-spec |
| 31 | 31 | # } |
| 32 | 32 | |
| 33 | 33 | |
| 34 | def _is_fsspec_url(url): | |
| 35 | return ( | |
| 36 | isinstance(url, str) | |
| 37 | and "://" in url | |
| 38 | and not url.startswith(("http://", "https://")) | |
| 39 | ) | |
| 40 | ||
| 41 | ||
| 34 | 42 | def _create_metadata(df): |
| 35 | 43 | """Create and encode geo metadata dict. |
| 36 | 44 | |
| 234 | 242 | "pyarrow.parquet", extra="pyarrow is required for Parquet support." |
| 235 | 243 | ) |
| 236 | 244 | |
| 245 | path = _expand_user(path) | |
| 237 | 246 | table = _geopandas_to_arrow(df, index=index) |
| 238 | 247 | parquet.write_table(table, path, compression=compression, **kwargs) |
| 239 | 248 | |
| 281 | 290 | if pyarrow.__version__ < LooseVersion("0.17.0"): |
| 282 | 291 | raise ImportError("pyarrow >= 0.17 required for Feather support") |
| 283 | 292 | |
| 293 | path = _expand_user(path) | |
| 284 | 294 | table = _geopandas_to_arrow(df, index=index) |
| 285 | 295 | feather.write_feather(table, path, compression=compression, **kwargs) |
| 286 | 296 | |
| 292 | 302 | df = table.to_pandas() |
| 293 | 303 | |
| 294 | 304 | metadata = table.schema.metadata |
| 295 | if b"geo" not in metadata: | |
| 305 | if metadata is None or b"geo" not in metadata: | |
| 296 | 306 | raise ValueError( |
| 297 | 307 | """Missing geo metadata in Parquet/Feather file. |
| 298 | 308 | Use pandas.read_parquet/read_feather() instead.""" |
| 338 | 348 | return GeoDataFrame(df, geometry=geometry) |
| 339 | 349 | |
| 340 | 350 | |
| 341 | def _read_parquet(path, columns=None, **kwargs): | |
| 351 | def _get_filesystem_path(path, filesystem=None, storage_options=None): | |
| 352 | """ | |
| 353 | Get the filesystem and path for a given filesystem and path. | |
| 354 | ||
| 355 | If the filesystem is not None then it's just returned as is. | |
| 356 | """ | |
| 357 | import pyarrow | |
| 358 | ||
| 359 | if ( | |
| 360 | isinstance(path, str) | |
| 361 | and storage_options is None | |
| 362 | and filesystem is None | |
| 363 | and LooseVersion(pyarrow.__version__) >= "5.0.0" | |
| 364 | ): | |
| 365 | # Use the native pyarrow filesystem if possible. | |
| 366 | try: | |
| 367 | from pyarrow.fs import FileSystem | |
| 368 | ||
| 369 | filesystem, path = FileSystem.from_uri(path) | |
| 370 | except Exception: | |
| 371 | # fallback to use get_handle / fsspec for filesystems | |
| 372 | # that pyarrow doesn't support | |
| 373 | pass | |
| 374 | ||
| 375 | if _is_fsspec_url(path) and filesystem is None: | |
| 376 | fsspec = import_optional_dependency( | |
| 377 | "fsspec", extra="fsspec is requred for 'storage_options'." | |
| 378 | ) | |
| 379 | filesystem, path = fsspec.core.url_to_fs(path, **(storage_options or {})) | |
| 380 | ||
| 381 | if filesystem is None and storage_options: | |
| 382 | raise ValueError( | |
| 383 | "Cannot provide 'storage_options' with non-fsspec path '{}'".format(path) | |
| 384 | ) | |
| 385 | ||
| 386 | return filesystem, path | |
| 387 | ||
| 388 | ||
| 389 | def _read_parquet(path, columns=None, storage_options=None, **kwargs): | |
| 342 | 390 | """ |
| 343 | 391 | Load a Parquet object from the file path, returning a GeoDataFrame. |
| 344 | 392 | |
| 365 | 413 | geometry read from the file will be set as the geometry column |
| 366 | 414 | of the returned GeoDataFrame. If no geometry columns are present, |
| 367 | 415 | a ``ValueError`` will be raised. |
| 416 | storage_options : dict, optional | |
| 417 | Extra options that make sense for a particular storage connection, e.g. host, | |
| 418 | port, username, password, etc. For HTTP(S) URLs the key-value pairs are | |
| 419 | forwarded to urllib as header options. For other URLs (e.g. starting with | |
| 420 | "s3://", and "gcs://") the key-value pairs are forwarded to fsspec. Please | |
| 421 | see fsspec and urllib for more details. | |
| 422 | ||
| 423 | When no storage options are provided and a filesystem is implemented by | |
| 424 | both ``pyarrow.fs`` and ``fsspec`` (e.g. "s3://") then the ``pyarrow.fs`` | |
| 425 | filesystem is preferred. Provide the instantiated fsspec filesystem using | |
| 426 | the ``filesystem`` keyword if you wish to use its implementation. | |
| 368 | 427 | **kwargs |
| 369 | 428 | Any additional kwargs passed to pyarrow.parquet.read_table(). |
| 370 | 429 | |
| 387 | 446 | parquet = import_optional_dependency( |
| 388 | 447 | "pyarrow.parquet", extra="pyarrow is required for Parquet support." |
| 389 | 448 | ) |
| 390 | ||
| 449 | # TODO(https://github.com/pandas-dev/pandas/pull/41194): see if pandas | |
| 450 | # adds filesystem as a keyword and match that. | |
| 451 | filesystem = kwargs.pop("filesystem", None) | |
| 452 | filesystem, path = _get_filesystem_path( | |
| 453 | path, filesystem=filesystem, storage_options=storage_options | |
| 454 | ) | |
| 455 | ||
| 456 | path = _expand_user(path) | |
| 391 | 457 | kwargs["use_pandas_metadata"] = True |
| 392 | table = parquet.read_table(path, columns=columns, **kwargs) | |
| 458 | table = parquet.read_table(path, columns=columns, filesystem=filesystem, **kwargs) | |
| 393 | 459 | |
| 394 | 460 | return _arrow_to_geopandas(table) |
| 395 | 461 | |
| 449 | 515 | if pyarrow.__version__ < LooseVersion("0.17.0"): |
| 450 | 516 | raise ImportError("pyarrow >= 0.17 required for Feather support") |
| 451 | 517 | |
| 518 | path = _expand_user(path) | |
| 452 | 519 | table = feather.read_table(path, columns=columns, **kwargs) |
| 453 | 520 | return _arrow_to_geopandas(table) |
| 0 | import os | |
| 0 | 1 | from distutils.version import LooseVersion |
| 1 | ||
| 2 | from pathlib import Path | |
| 2 | 3 | import warnings |
| 4 | ||
| 3 | 5 | import numpy as np |
| 4 | 6 | import pandas as pd |
| 5 | 7 | |
| 11 | 13 | import fiona |
| 12 | 14 | |
| 13 | 15 | fiona_import_error = None |
| 16 | ||
| 17 | # only try to import fiona.Env if the main fiona import succeeded (otherwise you | |
| 18 | # can get confusing "AttributeError: module 'fiona' has no attribute '_loading'" | |
| 19 | # / partially initialized module errors) | |
| 20 | try: | |
| 21 | from fiona import Env as fiona_env | |
| 22 | except ImportError: | |
| 23 | try: | |
| 24 | from fiona import drivers as fiona_env | |
| 25 | except ImportError: | |
| 26 | fiona_env = None | |
| 27 | ||
| 14 | 28 | except ImportError as err: |
| 15 | 29 | fiona = None |
| 16 | 30 | fiona_import_error = str(err) |
| 17 | 31 | |
| 18 | try: | |
| 19 | from fiona import Env as fiona_env | |
| 20 | except ImportError: | |
| 21 | try: | |
| 22 | from fiona import drivers as fiona_env | |
| 23 | except ImportError: | |
| 24 | fiona_env = None | |
| 25 | 32 | |
| 26 | 33 | from geopandas import GeoDataFrame, GeoSeries |
| 27 | 34 | |
| 34 | 41 | |
| 35 | 42 | _VALID_URLS = set(uses_relative + uses_netloc + uses_params) |
| 36 | 43 | _VALID_URLS.discard("") |
| 44 | ||
| 45 | _EXTENSION_TO_DRIVER = { | |
| 46 | ".bna": "BNA", | |
| 47 | ".dxf": "DXF", | |
| 48 | ".csv": "CSV", | |
| 49 | ".shp": "ESRI Shapefile", | |
| 50 | ".dbf": "ESRI Shapefile", | |
| 51 | ".json": "GeoJSON", | |
| 52 | ".geojson": "GeoJSON", | |
| 53 | ".geojsonl": "GeoJSONSeq", | |
| 54 | ".geojsons": "GeoJSONSeq", | |
| 55 | ".gpkg": "GPKG", | |
| 56 | ".gml": "GML", | |
| 57 | ".xml": "GML", | |
| 58 | ".gpx": "GPX", | |
| 59 | ".gtm": "GPSTrackMaker", | |
| 60 | ".gtz": "GPSTrackMaker", | |
| 61 | ".tab": "MapInfo File", | |
| 62 | ".mif": "MapInfo File", | |
| 63 | ".mid": "MapInfo File", | |
| 64 | ".dgn": "DGN", | |
| 65 | } | |
| 66 | ||
| 67 | ||
| 68 | def _expand_user(path): | |
| 69 | """Expand paths that use ~.""" | |
| 70 | if isinstance(path, str): | |
| 71 | path = os.path.expanduser(path) | |
| 72 | elif isinstance(path, Path): | |
| 73 | path = path.expanduser() | |
| 74 | return path | |
| 37 | 75 | |
| 38 | 76 | |
| 39 | 77 | def _check_fiona(func): |
| 77 | 115 | bbox : tuple | GeoDataFrame or GeoSeries | shapely Geometry, default None |
| 78 | 116 | Filter features by given bounding box, GeoSeries, GeoDataFrame or a |
| 79 | 117 | shapely geometry. CRS mis-matches are resolved if given a GeoSeries |
| 80 | or GeoDataFrame. Cannot be used with mask. | |
| 118 | or GeoDataFrame. Tuple is (minx, miny, maxx, maxy) to match the | |
| 119 | bounds property of shapely geometry objects. Cannot be used with mask. | |
| 81 | 120 | mask : dict | GeoDataFrame or GeoSeries | shapely Geometry, default None |
| 82 | 121 | Filter for features that intersect with the given dict-like geojson |
| 83 | 122 | geometry, GeoSeries, GeoDataFrame or shapely geometry. |
| 110 | 149 | |
| 111 | 150 | Reading only geometries intersecting ``bbox``: |
| 112 | 151 | |
| 113 | >>> df = geopandas.read_file("nybb.shp", bbox=(0, 10, 0, 20)) # doctest: +SKIP | |
| 152 | >>> df = geopandas.read_file("nybb.shp", bbox=(0, 0, 10, 20)) # doctest: +SKIP | |
| 114 | 153 | |
| 115 | 154 | Returns |
| 116 | 155 | ------- |
| 124 | 163 | by using the encoding keyword parameter, e.g. ``encoding='utf-8'``. |
| 125 | 164 | """ |
| 126 | 165 | _check_fiona("'read_file' function") |
| 166 | filename = _expand_user(filename) | |
| 167 | ||
| 127 | 168 | if _is_url(filename): |
| 128 | 169 | req = _urlopen(filename) |
| 129 | 170 | path_or_bytes = req.read() |
| 231 | 272 | return _to_file(*args, **kwargs) |
| 232 | 273 | |
| 233 | 274 | |
| 275 | def _detect_driver(path): | |
| 276 | """ | |
| 277 | Attempt to auto-detect driver based on the extension | |
| 278 | """ | |
| 279 | try: | |
| 280 | # in case the path is a file handle | |
| 281 | path = path.name | |
| 282 | except AttributeError: | |
| 283 | pass | |
| 284 | try: | |
| 285 | return _EXTENSION_TO_DRIVER[Path(path).suffix.lower()] | |
| 286 | except KeyError: | |
| 287 | # Assume it is a shapefile folder for now. In the future, | |
| 288 | # will likely raise an exception when the expected | |
| 289 | # folder writing behavior is more clearly defined. | |
| 290 | return "ESRI Shapefile" | |
| 291 | ||
| 292 | ||
| 234 | 293 | def _to_file( |
| 235 | 294 | df, |
| 236 | 295 | filename, |
| 237 | driver="ESRI Shapefile", | |
| 296 | driver=None, | |
| 238 | 297 | schema=None, |
| 239 | 298 | index=None, |
| 240 | 299 | mode="w", |
| 253 | 312 | df : GeoDataFrame to be written |
| 254 | 313 | filename : string |
| 255 | 314 | File path or file handle to write to. |
| 256 | driver : string, default 'ESRI Shapefile' | |
| 315 | driver : string, default None | |
| 257 | 316 | The OGR format driver used to write the vector file. |
| 317 | If not specified, it attempts to infer it from the file extension. | |
| 318 | If no extension is specified, it saves ESRI Shapefile to a folder. | |
| 258 | 319 | schema : dict, default None |
| 259 | 320 | If specified, the schema dictionary is passed to Fiona to |
| 260 | 321 | better control how the file is written. If None, GeoPandas |
| 291 | 352 | by using the encoding keyword parameter, e.g. ``encoding='utf-8'``. |
| 292 | 353 | """ |
| 293 | 354 | _check_fiona("'to_file' method") |
| 355 | filename = _expand_user(filename) | |
| 356 | ||
| 294 | 357 | if index is None: |
| 295 | 358 | # Determine if index attribute(s) should be saved to file |
| 296 | 359 | index = list(df.index.names) != [None] or type(df.index) not in ( |
| 306 | 369 | else: |
| 307 | 370 | crs = df.crs |
| 308 | 371 | |
| 372 | if driver is None: | |
| 373 | driver = _detect_driver(filename) | |
| 374 | ||
| 309 | 375 | if driver == "ESRI Shapefile" and any([len(c) > 10 for c in df.columns.tolist()]): |
| 310 | 376 | warnings.warn( |
| 311 | 377 | "Column names longer than 10 characters will be truncated when saved to " |
| 137 | 137 | PostGIS |
| 138 | 138 | |
| 139 | 139 | >>> from sqlalchemy import create_engine # doctest: +SKIP |
| 140 | >>> db_connection_url = "postgres://myusername:mypassword@myhost:5432/mydatabase" | |
| 140 | >>> db_connection_url = "postgresql://myusername:mypassword@myhost:5432/mydatabase" | |
| 141 | 141 | >>> con = create_engine(db_connection_url) # doctest: +SKIP |
| 142 | 142 | >>> sql = "SELECT geom, highway FROM roads" |
| 143 | 143 | >>> df = geopandas.read_postgis(sql, con) # doctest: +SKIP |
| 361 | 361 | -------- |
| 362 | 362 | |
| 363 | 363 | >>> from sqlalchemy import create_engine # doctest: +SKIP |
| 364 | >>> engine = create_engine("postgres://myusername:mypassword@myhost:5432\ | |
| 364 | >>> engine = create_engine("postgresql://myusername:mypassword@myhost:5432\ | |
| 365 | 365 | /mydatabase";) # doctest: +SKIP |
| 366 | 366 | >>> gdf.to_postgis("my_table", engine) # doctest: +SKIP |
| 367 | 367 | """ |
| 6 | 6 | from pandas import DataFrame, read_parquet as pd_read_parquet |
| 7 | 7 | from pandas.testing import assert_frame_equal |
| 8 | 8 | import numpy as np |
| 9 | from shapely.geometry import box | |
| 9 | 10 | |
| 10 | 11 | import geopandas |
| 11 | 12 | from geopandas import GeoDataFrame, read_file, read_parquet, read_feather |
| 15 | 16 | _create_metadata, |
| 16 | 17 | _decode_metadata, |
| 17 | 18 | _encode_metadata, |
| 19 | _get_filesystem_path, | |
| 18 | 20 | _validate_dataframe, |
| 19 | 21 | _validate_metadata, |
| 20 | 22 | METADATA_VERSION, |
| 337 | 339 | read_parquet(filename) |
| 338 | 340 | |
| 339 | 341 | |
| 342 | def test_parquet_missing_metadata2(tmpdir): | |
| 343 | """Missing geo metadata, such as from a parquet file created | |
| 344 | from a pyarrow Table (which will also not contain pandas metadata), | |
| 345 | will raise a ValueError. | |
| 346 | """ | |
| 347 | import pyarrow.parquet as pq | |
| 348 | ||
| 349 | table = pyarrow.table({"a": [1, 2, 3]}) | |
| 350 | filename = os.path.join(str(tmpdir), "test.pq") | |
| 351 | ||
| 352 | # use pyarrow.parquet write_table (no geo metadata, but also no pandas metadata) | |
| 353 | pq.write_table(table, filename) | |
| 354 | ||
| 355 | # missing metadata will raise ValueError | |
| 356 | with pytest.raises( | |
| 357 | ValueError, match="Missing geo metadata in Parquet/Feather file." | |
| 358 | ): | |
| 359 | read_parquet(filename) | |
| 360 | ||
| 361 | ||
| 340 | 362 | @pytest.mark.parametrize( |
| 341 | 363 | "geo_meta,error", |
| 342 | 364 | [ |
| 477 | 499 | ImportError, match="pyarrow >= 0.17 required for Feather support" |
| 478 | 500 | ): |
| 479 | 501 | df.to_feather(filename) |
| 502 | ||
| 503 | ||
| 504 | def test_fsspec_url(): | |
| 505 | fsspec = pytest.importorskip("fsspec") | |
| 506 | import fsspec.implementations.memory | |
| 507 | ||
| 508 | class MyMemoryFileSystem(fsspec.implementations.memory.MemoryFileSystem): | |
| 509 | # Simple fsspec filesystem that adds a required keyword. | |
| 510 | # Attempting to use this filesystem without the keyword will raise an exception. | |
| 511 | def __init__(self, is_set, *args, **kwargs): | |
| 512 | self.is_set = is_set | |
| 513 | super().__init__(*args, **kwargs) | |
| 514 | ||
| 515 | fsspec.register_implementation("memory", MyMemoryFileSystem, clobber=True) | |
| 516 | memfs = MyMemoryFileSystem(is_set=True) | |
| 517 | ||
| 518 | test_dataset = "naturalearth_lowres" | |
| 519 | df = read_file(get_path(test_dataset)) | |
| 520 | ||
| 521 | with memfs.open("data.parquet", "wb") as f: | |
| 522 | df.to_parquet(f) | |
| 523 | ||
| 524 | result = read_parquet("memory://data.parquet", storage_options=dict(is_set=True)) | |
| 525 | assert_geodataframe_equal(result, df) | |
| 526 | ||
| 527 | result = read_parquet("memory://data.parquet", filesystem=memfs) | |
| 528 | assert_geodataframe_equal(result, df) | |
| 529 | ||
| 530 | ||
| 531 | def test_non_fsspec_url_with_storage_options_raises(): | |
| 532 | with pytest.raises(ValueError, match="storage_options"): | |
| 533 | test_dataset = "naturalearth_lowres" | |
| 534 | read_parquet(get_path(test_dataset), storage_options={"foo": "bar"}) | |
| 535 | ||
| 536 | ||
| 537 | @pytest.mark.skipif( | |
| 538 | pyarrow.__version__ < LooseVersion("5.0.0"), | |
| 539 | reason="pyarrow.fs requires pyarrow>=5.0.0", | |
| 540 | ) | |
| 541 | def test_prefers_pyarrow_fs(): | |
| 542 | filesystem, _ = _get_filesystem_path("file:///data.parquet") | |
| 543 | assert isinstance(filesystem, pyarrow.fs.LocalFileSystem) | |
| 544 | ||
| 545 | ||
| 546 | def test_write_read_parquet_expand_user(): | |
| 547 | gdf = geopandas.GeoDataFrame(geometry=[box(0, 0, 10, 10)], crs="epsg:4326") | |
| 548 | test_file = "~/test_file.parquet" | |
| 549 | gdf.to_parquet(test_file) | |
| 550 | pq_df = geopandas.read_parquet(test_file) | |
| 551 | assert_geodataframe_equal(gdf, pq_df, check_crs=True) | |
| 552 | os.remove(os.path.expanduser(test_file)) | |
| 553 | ||
| 554 | ||
| 555 | def test_write_read_feather_expand_user(): | |
| 556 | gdf = geopandas.GeoDataFrame(geometry=[box(0, 0, 10, 10)], crs="epsg:4326") | |
| 557 | test_file = "~/test_file.feather" | |
| 558 | gdf.to_feather(test_file) | |
| 559 | f_df = geopandas.read_feather(test_file) | |
| 560 | assert_geodataframe_equal(gdf, f_df, check_crs=True) | |
| 561 | os.remove(os.path.expanduser(test_file)) | |
| 12 | 12 | |
| 13 | 13 | import geopandas |
| 14 | 14 | from geopandas import GeoDataFrame, read_file |
| 15 | from geopandas.io.file import fiona_env | |
| 15 | from geopandas.io.file import fiona_env, _detect_driver, _EXTENSION_TO_DRIVER | |
| 16 | 16 | |
| 17 | 17 | from geopandas.testing import assert_geodataframe_equal, assert_geoseries_equal |
| 18 | 18 | from geopandas.tests.util import PACKAGE_DIR, validate_boro_df |
| 60 | 60 | # to_file tests |
| 61 | 61 | # ----------------------------------------------------------------------------- |
| 62 | 62 | |
| 63 | driver_ext_pairs = [("ESRI Shapefile", "shp"), ("GeoJSON", "geojson"), ("GPKG", "gpkg")] | |
| 63 | driver_ext_pairs = [ | |
| 64 | ("ESRI Shapefile", ".shp"), | |
| 65 | ("GeoJSON", ".geojson"), | |
| 66 | ("GPKG", ".gpkg"), | |
| 67 | (None, ".shp"), | |
| 68 | (None, ""), | |
| 69 | (None, ".geojson"), | |
| 70 | (None, ".gpkg"), | |
| 71 | ] | |
| 72 | ||
| 73 | ||
| 74 | def assert_correct_driver(file_path, ext): | |
| 75 | # check the expected driver | |
| 76 | expected_driver = "ESRI Shapefile" if ext == "" else _EXTENSION_TO_DRIVER[ext] | |
| 77 | with fiona.open(str(file_path)) as fds: | |
| 78 | assert fds.driver == expected_driver | |
| 64 | 79 | |
| 65 | 80 | |
| 66 | 81 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) |
| 67 | 82 | def test_to_file(tmpdir, df_nybb, df_null, driver, ext): |
| 68 | """ Test to_file and from_file """ | |
| 83 | """Test to_file and from_file""" | |
| 69 | 84 | tempfilename = os.path.join(str(tmpdir), "boros." + ext) |
| 70 | 85 | df_nybb.to_file(tempfilename, driver=driver) |
| 71 | 86 | # Read layer back in |
| 75 | 90 | assert np.alltrue(df["BoroName"].values == df_nybb["BoroName"]) |
| 76 | 91 | |
| 77 | 92 | # Write layer with null geometry out to file |
| 78 | tempfilename = os.path.join(str(tmpdir), "null_geom." + ext) | |
| 93 | tempfilename = os.path.join(str(tmpdir), "null_geom" + ext) | |
| 79 | 94 | df_null.to_file(tempfilename, driver=driver) |
| 80 | 95 | # Read layer back in |
| 81 | 96 | df = GeoDataFrame.from_file(tempfilename) |
| 82 | 97 | assert "geometry" in df |
| 83 | 98 | assert len(df) == 2 |
| 84 | 99 | assert np.alltrue(df["Name"].values == df_null["Name"]) |
| 100 | # check the expected driver | |
| 101 | assert_correct_driver(tempfilename, ext) | |
| 85 | 102 | |
| 86 | 103 | |
| 87 | 104 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) |
| 88 | 105 | def test_to_file_pathlib(tmpdir, df_nybb, df_null, driver, ext): |
| 89 | """ Test to_file and from_file """ | |
| 106 | """Test to_file and from_file""" | |
| 90 | 107 | temppath = pathlib.Path(os.path.join(str(tmpdir), "boros." + ext)) |
| 91 | 108 | df_nybb.to_file(temppath, driver=driver) |
| 92 | 109 | # Read layer back in |
| 94 | 111 | assert "geometry" in df |
| 95 | 112 | assert len(df) == 5 |
| 96 | 113 | assert np.alltrue(df["BoroName"].values == df_nybb["BoroName"]) |
| 114 | # check the expected driver | |
| 115 | assert_correct_driver(temppath, ext) | |
| 97 | 116 | |
| 98 | 117 | |
| 99 | 118 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) |
| 105 | 124 | "a": [1, 2, 3], |
| 106 | 125 | "b": [True, False, True], |
| 107 | 126 | "geometry": [Point(0, 0), Point(1, 1), Point(2, 2)], |
| 108 | } | |
| 127 | }, | |
| 128 | crs=4326, | |
| 109 | 129 | ) |
| 110 | 130 | |
| 111 | 131 | df.to_file(tempfilename, driver=driver) |
| 112 | 132 | result = read_file(tempfilename) |
| 113 | if driver == "GeoJSON": | |
| 114 | # geojson by default assumes epsg:4326 | |
| 115 | result.crs = None | |
| 116 | if driver == "ESRI Shapefile": | |
| 133 | if ext in (".shp", ""): | |
| 117 | 134 | # Shapefile does not support boolean, so is read back as int |
| 118 | 135 | df["b"] = df["b"].astype("int64") |
| 119 | 136 | assert_geodataframe_equal(result, df) |
| 137 | # check the expected driver | |
| 138 | assert_correct_driver(tempfilename, ext) | |
| 120 | 139 | |
| 121 | 140 | |
| 122 | 141 | def test_to_file_datetime(tmpdir): |
| 124 | 143 | tempfilename = os.path.join(str(tmpdir), "test_datetime.gpkg") |
| 125 | 144 | point = Point(0, 0) |
| 126 | 145 | now = datetime.datetime.now() |
| 127 | df = GeoDataFrame({"a": [1, 2], "b": [now, now]}, geometry=[point, point], crs={}) | |
| 146 | df = GeoDataFrame({"a": [1, 2], "b": [now, now]}, geometry=[point, point], crs=4326) | |
| 128 | 147 | df.to_file(tempfilename, driver="GPKG") |
| 129 | 148 | df_read = read_file(tempfilename) |
| 130 | 149 | assert_geoseries_equal(df.geometry, df_read.geometry) |
| 134 | 153 | def test_to_file_with_point_z(tmpdir, ext, driver): |
| 135 | 154 | """Test that 3D geometries are retained in writes (GH #612).""" |
| 136 | 155 | |
| 137 | tempfilename = os.path.join(str(tmpdir), "test_3Dpoint." + ext) | |
| 156 | tempfilename = os.path.join(str(tmpdir), "test_3Dpoint" + ext) | |
| 138 | 157 | point3d = Point(0, 0, 500) |
| 139 | 158 | point2d = Point(1, 1) |
| 140 | 159 | df = GeoDataFrame({"a": [1, 2]}, geometry=[point3d, point2d], crs=_CRS) |
| 141 | 160 | df.to_file(tempfilename, driver=driver) |
| 142 | 161 | df_read = GeoDataFrame.from_file(tempfilename) |
| 143 | 162 | assert_geoseries_equal(df.geometry, df_read.geometry) |
| 163 | # check the expected driver | |
| 164 | assert_correct_driver(tempfilename, ext) | |
| 144 | 165 | |
| 145 | 166 | |
| 146 | 167 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) |
| 147 | 168 | def test_to_file_with_poly_z(tmpdir, ext, driver): |
| 148 | 169 | """Test that 3D geometries are retained in writes (GH #612).""" |
| 149 | 170 | |
| 150 | tempfilename = os.path.join(str(tmpdir), "test_3Dpoly." + ext) | |
| 171 | tempfilename = os.path.join(str(tmpdir), "test_3Dpoly" + ext) | |
| 151 | 172 | poly3d = Polygon([[0, 0, 5], [0, 1, 5], [1, 1, 5], [1, 0, 5]]) |
| 152 | 173 | poly2d = Polygon([[0, 0], [0, 1], [1, 1], [1, 0]]) |
| 153 | 174 | df = GeoDataFrame({"a": [1, 2]}, geometry=[poly3d, poly2d], crs=_CRS) |
| 154 | 175 | df.to_file(tempfilename, driver=driver) |
| 155 | 176 | df_read = GeoDataFrame.from_file(tempfilename) |
| 156 | 177 | assert_geoseries_equal(df.geometry, df_read.geometry) |
| 178 | # check the expected driver | |
| 179 | assert_correct_driver(tempfilename, ext) | |
| 157 | 180 | |
| 158 | 181 | |
| 159 | 182 | def test_to_file_types(tmpdir, df_points): |
| 160 | """ Test various integer type columns (GH#93) """ | |
| 183 | """Test various integer type columns (GH#93)""" | |
| 161 | 184 | tempfilename = os.path.join(str(tmpdir), "int.shp") |
| 162 | 185 | int_types = [ |
| 163 | 186 | np.int8, |
| 246 | 269 | |
| 247 | 270 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) |
| 248 | 271 | def test_append_file(tmpdir, df_nybb, df_null, driver, ext): |
| 249 | """ Test to_file with append mode and from_file """ | |
| 272 | """Test to_file with append mode and from_file""" | |
| 250 | 273 | from fiona import supported_drivers |
| 251 | 274 | |
| 275 | tempfilename = os.path.join(str(tmpdir), "boros" + ext) | |
| 276 | driver = driver if driver else _detect_driver(tempfilename) | |
| 252 | 277 | if "a" not in supported_drivers[driver]: |
| 253 | 278 | return None |
| 254 | 279 | |
| 255 | tempfilename = os.path.join(str(tmpdir), "boros." + ext) | |
| 256 | 280 | df_nybb.to_file(tempfilename, driver=driver) |
| 257 | 281 | df_nybb.to_file(tempfilename, mode="a", driver=driver) |
| 258 | 282 | # Read layer back in |
| 263 | 287 | assert_geodataframe_equal(df, expected, check_less_precise=True) |
| 264 | 288 | |
| 265 | 289 | # Write layer with null geometry out to file |
| 266 | tempfilename = os.path.join(str(tmpdir), "null_geom." + ext) | |
| 290 | tempfilename = os.path.join(str(tmpdir), "null_geom" + ext) | |
| 267 | 291 | df_null.to_file(tempfilename, driver=driver) |
| 268 | 292 | df_null.to_file(tempfilename, mode="a", driver=driver) |
| 269 | 293 | # Read layer back in |
| 272 | 296 | assert len(df) == (2 * 2) |
| 273 | 297 | expected = pd.concat([df_null] * 2, ignore_index=True) |
| 274 | 298 | assert_geodataframe_equal(df, expected, check_less_precise=True) |
| 299 | ||
| 300 | ||
| 301 | @pytest.mark.parametrize("driver,ext", driver_ext_pairs) | |
| 302 | def test_empty_crs(tmpdir, driver, ext): | |
| 303 | """Test handling of undefined CRS with GPKG driver (GH #1975).""" | |
| 304 | if ext == ".gpkg": | |
| 305 | pytest.xfail("GPKG is read with Undefined geographic SRS.") | |
| 306 | ||
| 307 | tempfilename = os.path.join(str(tmpdir), "boros" + ext) | |
| 308 | df = GeoDataFrame( | |
| 309 | { | |
| 310 | "a": [1, 2, 3], | |
| 311 | "geometry": [Point(0, 0), Point(1, 1), Point(2, 2)], | |
| 312 | }, | |
| 313 | ) | |
| 314 | ||
| 315 | df.to_file(tempfilename, driver=driver) | |
| 316 | result = read_file(tempfilename) | |
| 317 | ||
| 318 | if ext == ".geojson": | |
| 319 | # geojson by default assumes epsg:4326 | |
| 320 | df.crs = "EPSG:4326" | |
| 321 | ||
| 322 | assert_geodataframe_equal(result, df) | |
| 275 | 323 | |
| 276 | 324 | |
| 277 | 325 | # ----------------------------------------------------------------------------- |
| 389 | 437 | gdf = read_file(path) |
| 390 | 438 | assert isinstance(gdf, geopandas.GeoDataFrame) |
| 391 | 439 | |
| 392 | # Check that it can sucessfully add a zip scheme to a path that already has a scheme | |
| 440 | # Check that it can successfully add a zip scheme to a path that already has a | |
| 441 | # scheme | |
| 393 | 442 | gdf = read_file("file+file://" + path) |
| 394 | 443 | assert isinstance(gdf, geopandas.GeoDataFrame) |
| 395 | 444 | |
| 406 | 455 | assert isinstance(gdf, geopandas.GeoDataFrame) |
| 407 | 456 | |
| 408 | 457 | |
| 409 | def test_read_file_filtered(df_nybb): | |
| 410 | full_df_shape = df_nybb.shape | |
| 458 | def test_read_file_filtered__bbox(df_nybb): | |
| 411 | 459 | nybb_filename = geopandas.datasets.get_path("nybb") |
| 412 | 460 | bbox = ( |
| 413 | 461 | 1031051.7879884212, |
| 416 | 464 | 244317.30894023244, |
| 417 | 465 | ) |
| 418 | 466 | filtered_df = read_file(nybb_filename, bbox=bbox) |
| 419 | filtered_df_shape = filtered_df.shape | |
| 420 | assert full_df_shape != filtered_df_shape | |
| 421 | assert filtered_df_shape == (2, 5) | |
| 467 | expected = df_nybb[df_nybb["BoroName"].isin(["Bronx", "Queens"])] | |
| 468 | assert_geodataframe_equal(filtered_df, expected.reset_index(drop=True)) | |
| 469 | ||
| 470 | ||
| 471 | def test_read_file_filtered__bbox__polygon(df_nybb): | |
| 472 | nybb_filename = geopandas.datasets.get_path("nybb") | |
| 473 | bbox = box( | |
| 474 | 1031051.7879884212, 224272.49231459625, 1047224.3104931959, 244317.30894023244 | |
| 475 | ) | |
| 476 | filtered_df = read_file(nybb_filename, bbox=bbox) | |
| 477 | expected = df_nybb[df_nybb["BoroName"].isin(["Bronx", "Queens"])] | |
| 478 | assert_geodataframe_equal(filtered_df, expected.reset_index(drop=True)) | |
| 422 | 479 | |
| 423 | 480 | |
| 424 | 481 | def test_read_file_filtered__rows(df_nybb): |
| 425 | full_df_shape = df_nybb.shape | |
| 426 | 482 | nybb_filename = geopandas.datasets.get_path("nybb") |
| 427 | 483 | filtered_df = read_file(nybb_filename, rows=1) |
| 428 | filtered_df_shape = filtered_df.shape | |
| 429 | assert full_df_shape != filtered_df_shape | |
| 430 | assert filtered_df_shape == (1, 5) | |
| 431 | ||
| 432 | ||
| 484 | assert_geodataframe_equal(filtered_df, df_nybb.iloc[[0], :]) | |
| 485 | ||
| 486 | ||
| 487 | def test_read_file_filtered__rows_slice(df_nybb): | |
| 488 | nybb_filename = geopandas.datasets.get_path("nybb") | |
| 489 | filtered_df = read_file(nybb_filename, rows=slice(1, 3)) | |
| 490 | assert_geodataframe_equal(filtered_df, df_nybb.iloc[1:3, :].reset_index(drop=True)) | |
| 491 | ||
| 492 | ||
| 493 | @pytest.mark.filterwarnings( | |
| 494 | "ignore:Layer does not support OLC_FASTFEATURECOUNT:RuntimeWarning" | |
| 495 | ) # for the slice with -1 | |
| 433 | 496 | def test_read_file_filtered__rows_bbox(df_nybb): |
| 434 | full_df_shape = df_nybb.shape | |
| 435 | 497 | nybb_filename = geopandas.datasets.get_path("nybb") |
| 436 | 498 | bbox = ( |
| 437 | 499 | 1031051.7879884212, |
| 439 | 501 | 1047224.3104931959, |
| 440 | 502 | 244317.30894023244, |
| 441 | 503 | ) |
| 504 | # combination bbox and rows (rows slice applied after bbox filtering!) | |
| 505 | filtered_df = read_file(nybb_filename, bbox=bbox, rows=slice(4, None)) | |
| 506 | assert filtered_df.empty | |
| 442 | 507 | filtered_df = read_file(nybb_filename, bbox=bbox, rows=slice(-1, None)) |
| 443 | filtered_df_shape = filtered_df.shape | |
| 444 | assert full_df_shape != filtered_df_shape | |
| 445 | assert filtered_df_shape == (1, 5) | |
| 446 | ||
| 447 | ||
| 448 | def test_read_file_filtered__rows_bbox__polygon(df_nybb): | |
| 449 | full_df_shape = df_nybb.shape | |
| 450 | nybb_filename = geopandas.datasets.get_path("nybb") | |
| 451 | bbox = box( | |
| 452 | 1031051.7879884212, 224272.49231459625, 1047224.3104931959, 244317.30894023244 | |
| 453 | ) | |
| 454 | filtered_df = read_file(nybb_filename, bbox=bbox, rows=slice(-1, None)) | |
| 455 | filtered_df_shape = filtered_df.shape | |
| 456 | assert full_df_shape != filtered_df_shape | |
| 457 | assert filtered_df_shape == (1, 5) | |
| 508 | assert_geodataframe_equal(filtered_df, df_nybb.iloc[4:, :].reset_index(drop=True)) | |
| 458 | 509 | |
| 459 | 510 | |
| 460 | 511 | def test_read_file_filtered_rows_invalid(): |
| 778 | 829 | # named DatetimeIndex |
| 779 | 830 | df.index.name = "datetime" |
| 780 | 831 | do_checks(df, index_is_used=True) |
| 832 | ||
| 833 | ||
| 834 | def test_to_file__undetermined_driver(tmp_path, df_nybb): | |
| 835 | shpdir = tmp_path / "boros.invalid" | |
| 836 | df_nybb.to_file(shpdir) | |
| 837 | assert shpdir.is_dir() | |
| 838 | assert list(shpdir.glob("*.shp")) | |
| 839 | ||
| 840 | ||
| 841 | @pytest.mark.parametrize( | |
| 842 | "test_file", [(pathlib.Path("~/test_file.geojson")), "~/test_file.geojson"] | |
| 843 | ) | |
| 844 | def test_write_read_file(test_file): | |
| 845 | gdf = geopandas.GeoDataFrame(geometry=[box(0, 0, 10, 10)], crs=_CRS) | |
| 846 | gdf.to_file(test_file, driver="GeoJSON") | |
| 847 | df_json = geopandas.read_file(test_file) | |
| 848 | assert_geodataframe_equal(gdf, df_json, check_crs=True) | |
| 849 | os.remove(os.path.expanduser(test_file)) | |
| 63 | 63 | |
| 64 | 64 | try: |
| 65 | 65 | con = sqlalchemy.create_engine( |
| 66 | URL( | |
| 66 | URL.create( | |
| 67 | 67 | drivername="postgresql+psycopg2", |
| 68 | 68 | username=user, |
| 69 | 69 | database=dbname, |
| 113 | 113 | def drop_table_if_exists(conn_or_engine, table): |
| 114 | 114 | sqlalchemy = pytest.importorskip("sqlalchemy") |
| 115 | 115 | |
| 116 | if conn_or_engine.dialect.has_table(conn_or_engine, table): | |
| 116 | if sqlalchemy.inspect(conn_or_engine).has_table(table): | |
| 117 | 117 | metadata = sqlalchemy.MetaData(conn_or_engine) |
| 118 | 118 | metadata.reflect() |
| 119 | 119 | table = metadata.tables.get(table) |
| 1 | 1 | |
| 2 | 2 | import numpy as np |
| 3 | 3 | import pandas as pd |
| 4 | from pandas.plotting import PlotAccessor | |
| 4 | 5 | |
| 5 | 6 | import geopandas |
| 6 | 7 | |
| 7 | 8 | from distutils.version import LooseVersion |
| 9 | ||
| 10 | from ._decorator import doc | |
| 8 | 11 | |
| 9 | 12 | |
| 10 | 13 | def deprecated(new): |
| 69 | 72 | mpl = matplotlib.__version__ |
| 70 | 73 | if mpl >= LooseVersion("3.4") or (mpl > LooseVersion("3.3.2") and "+" in mpl): |
| 71 | 74 | # alpha is supported as array argument with matplotlib 3.4+ |
| 72 | scalar_kwargs = ["marker"] | |
| 75 | scalar_kwargs = ["marker", "path_effects"] | |
| 73 | 76 | else: |
| 74 | scalar_kwargs = ["marker", "alpha"] | |
| 77 | scalar_kwargs = ["marker", "alpha", "path_effects"] | |
| 75 | 78 | |
| 76 | 79 | for att, value in kwargs.items(): |
| 77 | 80 | if "color" in att: # color(s), edgecolor(s), facecolor(s) |
| 722 | 725 | |
| 723 | 726 | nan_idx = np.asarray(pd.isna(values), dtype="bool") |
| 724 | 727 | |
| 725 | # Define `values` as a Series | |
| 726 | if categorical: | |
| 727 | if cmap is None: | |
| 728 | cmap = "tab10" | |
| 729 | ||
| 730 | cat = pd.Categorical(values, categories=categories) | |
| 731 | categories = list(cat.categories) | |
| 732 | ||
| 733 | # values missing in the Categorical but not in original values | |
| 734 | missing = list(np.unique(values[~nan_idx & cat.isna()])) | |
| 735 | if missing: | |
| 736 | raise ValueError( | |
| 737 | "Column contains values not listed in categories. " | |
| 738 | "Missing categories: {}.".format(missing) | |
| 739 | ) | |
| 740 | ||
| 741 | values = cat.codes[~nan_idx] | |
| 742 | vmin = 0 if vmin is None else vmin | |
| 743 | vmax = len(categories) - 1 if vmax is None else vmax | |
| 744 | ||
| 745 | 728 | if scheme is not None: |
| 729 | mc_err = ( | |
| 730 | "The 'mapclassify' package (>= 2.4.0) is " | |
| 731 | "required to use the 'scheme' keyword." | |
| 732 | ) | |
| 733 | try: | |
| 734 | import mapclassify | |
| 735 | ||
| 736 | except ImportError: | |
| 737 | raise ImportError(mc_err) | |
| 738 | ||
| 739 | if mapclassify.__version__ < LooseVersion("2.4.0"): | |
| 740 | raise ImportError(mc_err) | |
| 741 | ||
| 746 | 742 | if classification_kwds is None: |
| 747 | 743 | classification_kwds = {} |
| 748 | 744 | if "k" not in classification_kwds: |
| 749 | 745 | classification_kwds["k"] = k |
| 750 | 746 | |
| 751 | binning = _mapclassify_choro(values[~nan_idx], scheme, **classification_kwds) | |
| 747 | binning = mapclassify.classify( | |
| 748 | np.asarray(values[~nan_idx]), scheme, **classification_kwds | |
| 749 | ) | |
| 752 | 750 | # set categorical to True for creating the legend |
| 753 | 751 | categorical = True |
| 754 | 752 | if legend_kwds is not None and "labels" in legend_kwds: |
| 760 | 758 | ) |
| 761 | 759 | ) |
| 762 | 760 | else: |
| 763 | categories = list(legend_kwds.pop("labels")) | |
| 761 | labels = list(legend_kwds.pop("labels")) | |
| 764 | 762 | else: |
| 765 | 763 | fmt = "{:.2f}" |
| 766 | 764 | if legend_kwds is not None and "fmt" in legend_kwds: |
| 767 | 765 | fmt = legend_kwds.pop("fmt") |
| 768 | 766 | |
| 769 | categories = binning.get_legend_classes(fmt) | |
| 767 | labels = binning.get_legend_classes(fmt) | |
| 770 | 768 | if legend_kwds is not None: |
| 771 | 769 | show_interval = legend_kwds.pop("interval", False) |
| 772 | 770 | else: |
| 773 | 771 | show_interval = False |
| 774 | 772 | if not show_interval: |
| 775 | categories = [c[1:-1] for c in categories] | |
| 776 | values = np.array(binning.yb) | |
| 773 | labels = [c[1:-1] for c in labels] | |
| 774 | ||
| 775 | values = pd.Categorical([np.nan] * len(values), categories=labels, ordered=True) | |
| 776 | values[~nan_idx] = pd.Categorical.from_codes( | |
| 777 | binning.yb, categories=labels, ordered=True | |
| 778 | ) | |
| 779 | if cmap is None: | |
| 780 | cmap = "viridis" | |
| 781 | ||
| 782 | # Define `values` as a Series | |
| 783 | if categorical: | |
| 784 | if cmap is None: | |
| 785 | cmap = "tab10" | |
| 786 | ||
| 787 | cat = pd.Categorical(values, categories=categories) | |
| 788 | categories = list(cat.categories) | |
| 789 | ||
| 790 | # values missing in the Categorical but not in original values | |
| 791 | missing = list(np.unique(values[~nan_idx & cat.isna()])) | |
| 792 | if missing: | |
| 793 | raise ValueError( | |
| 794 | "Column contains values not listed in categories. " | |
| 795 | "Missing categories: {}.".format(missing) | |
| 796 | ) | |
| 797 | ||
| 798 | values = cat.codes[~nan_idx] | |
| 799 | vmin = 0 if vmin is None else vmin | |
| 800 | vmax = len(categories) - 1 if vmax is None else vmax | |
| 777 | 801 | |
| 778 | 802 | # fill values with placeholder where were NaNs originally to map them properly |
| 779 | 803 | # (after removing them in categorical or scheme) |
| 902 | 926 | else: |
| 903 | 927 | legend_kwds.setdefault("ax", ax) |
| 904 | 928 | |
| 905 | n_cmap.set_array([]) | |
| 929 | n_cmap.set_array(np.array([])) | |
| 906 | 930 | ax.get_figure().colorbar(n_cmap, **legend_kwds) |
| 907 | 931 | |
| 908 | 932 | plt.draw() |
| 909 | 933 | return ax |
| 910 | 934 | |
| 911 | 935 | |
| 912 | if geopandas._compat.PANDAS_GE_025: | |
| 913 | from pandas.plotting import PlotAccessor | |
| 914 | ||
| 915 | class GeoplotAccessor(PlotAccessor): | |
| 916 | ||
| 917 | __doc__ = plot_dataframe.__doc__ | |
| 918 | _pandas_kinds = PlotAccessor._all_kinds | |
| 919 | ||
| 920 | def __call__(self, *args, **kwargs): | |
| 921 | data = self._parent.copy() | |
| 922 | kind = kwargs.pop("kind", "geo") | |
| 923 | if kind == "geo": | |
| 924 | return plot_dataframe(data, *args, **kwargs) | |
| 925 | if kind in self._pandas_kinds: | |
| 926 | # Access pandas plots | |
| 927 | return PlotAccessor(data)(kind=kind, **kwargs) | |
| 928 | else: | |
| 929 | # raise error | |
| 930 | raise ValueError(f"{kind} is not a valid plot kind") | |
| 931 | ||
| 932 | def geo(self, *args, **kwargs): | |
| 933 | return self(kind="geo", *args, **kwargs) | |
| 934 | ||
| 935 | ||
| 936 | def _mapclassify_choro(values, scheme, **classification_kwds): | |
| 937 | """ | |
| 938 | Wrapper for choropleth schemes from mapclassify for use with plot_dataframe | |
| 939 | ||
| 940 | Parameters | |
| 941 | ---------- | |
| 942 | values | |
| 943 | Series to be plotted | |
| 944 | scheme : str | |
| 945 | One of mapclassify classification schemes | |
| 946 | Options are BoxPlot, EqualInterval, FisherJenks, | |
| 947 | FisherJenksSampled, HeadTailBreaks, JenksCaspall, | |
| 948 | JenksCaspallForced, JenksCaspallSampled, MaxP, | |
| 949 | MaximumBreaks, NaturalBreaks, Quantiles, Percentiles, StdMean, | |
| 950 | UserDefined | |
| 951 | ||
| 952 | **classification_kwds : dict | |
| 953 | Keyword arguments for classification scheme | |
| 954 | For details see mapclassify documentation: | |
| 955 | https://pysal.org/mapclassify/api.html | |
| 956 | ||
| 957 | Returns | |
| 958 | ------- | |
| 959 | binning | |
| 960 | Binning objects that holds the Series with values replaced with | |
| 961 | class identifier and the bins. | |
| 962 | """ | |
| 963 | try: | |
| 964 | import mapclassify.classifiers as classifiers | |
| 965 | ||
| 966 | except ImportError: | |
| 967 | raise ImportError( | |
| 968 | "The 'mapclassify' >= 2.2.0 package is required to use the 'scheme' keyword" | |
| 969 | ) | |
| 970 | from mapclassify import __version__ as mc_version | |
| 971 | ||
| 972 | if mc_version < LooseVersion("2.2.0"): | |
| 973 | raise ImportError( | |
| 974 | "The 'mapclassify' >= 2.2.0 package is required to " | |
| 975 | "use the 'scheme' keyword" | |
| 976 | ) | |
| 977 | schemes = {} | |
| 978 | for classifier in classifiers.CLASSIFIERS: | |
| 979 | schemes[classifier.lower()] = getattr(classifiers, classifier) | |
| 980 | ||
| 981 | scheme = scheme.lower() | |
| 982 | ||
| 983 | # mapclassify < 2.1 cleaned up the scheme names (removing underscores) | |
| 984 | # trying both to keep compatibility with older versions and provide | |
| 985 | # compatibility with newer versions of mapclassify | |
| 986 | oldnew = { | |
| 987 | "Box_Plot": "BoxPlot", | |
| 988 | "Equal_Interval": "EqualInterval", | |
| 989 | "Fisher_Jenks": "FisherJenks", | |
| 990 | "Fisher_Jenks_Sampled": "FisherJenksSampled", | |
| 991 | "HeadTail_Breaks": "HeadTailBreaks", | |
| 992 | "Jenks_Caspall": "JenksCaspall", | |
| 993 | "Jenks_Caspall_Forced": "JenksCaspallForced", | |
| 994 | "Jenks_Caspall_Sampled": "JenksCaspallSampled", | |
| 995 | "Max_P_Plassifier": "MaxP", | |
| 996 | "Maximum_Breaks": "MaximumBreaks", | |
| 997 | "Natural_Breaks": "NaturalBreaks", | |
| 998 | "Std_Mean": "StdMean", | |
| 999 | "User_Defined": "UserDefined", | |
| 1000 | } | |
| 1001 | scheme_names_mapping = {} | |
| 1002 | scheme_names_mapping.update( | |
| 1003 | {old.lower(): new.lower() for old, new in oldnew.items()} | |
| 1004 | ) | |
| 1005 | scheme_names_mapping.update( | |
| 1006 | {new.lower(): old.lower() for old, new in oldnew.items()} | |
| 1007 | ) | |
| 1008 | ||
| 1009 | try: | |
| 1010 | scheme_class = schemes[scheme] | |
| 1011 | except KeyError: | |
| 1012 | scheme = scheme_names_mapping.get(scheme, scheme) | |
| 1013 | try: | |
| 1014 | scheme_class = schemes[scheme] | |
| 1015 | except KeyError: | |
| 1016 | raise ValueError( | |
| 1017 | "Invalid scheme. Scheme must be in the set: %r" % schemes.keys() | |
| 1018 | ) | |
| 1019 | ||
| 1020 | if classification_kwds["k"] is not None: | |
| 1021 | from inspect import getfullargspec as getspec | |
| 1022 | ||
| 1023 | spec = getspec(scheme_class.__init__) | |
| 1024 | if "k" not in spec.args: | |
| 1025 | del classification_kwds["k"] | |
| 1026 | try: | |
| 1027 | binning = scheme_class(np.asarray(values), **classification_kwds) | |
| 1028 | except TypeError: | |
| 1029 | raise TypeError("Invalid keyword argument for %r " % scheme) | |
| 1030 | return binning | |
| 936 | @doc(plot_dataframe) | |
| 937 | class GeoplotAccessor(PlotAccessor): | |
| 938 | ||
| 939 | _pandas_kinds = PlotAccessor._all_kinds | |
| 940 | ||
| 941 | def __call__(self, *args, **kwargs): | |
| 942 | data = self._parent.copy() | |
| 943 | kind = kwargs.pop("kind", "geo") | |
| 944 | if kind == "geo": | |
| 945 | return plot_dataframe(data, *args, **kwargs) | |
| 946 | if kind in self._pandas_kinds: | |
| 947 | # Access pandas plots | |
| 948 | return PlotAccessor(data)(kind=kind, **kwargs) | |
| 949 | else: | |
| 950 | # raise error | |
| 951 | raise ValueError(f"{kind} is not a valid plot kind") | |
| 952 | ||
| 953 | def geo(self, *args, **kwargs): | |
| 954 | return self(kind="geo", *args, **kwargs) | |
| 0 | from textwrap import dedent | |
| 1 | 0 | import warnings |
| 2 | 1 | |
| 3 | 2 | from shapely.geometry.base import BaseGeometry |
| 5 | 4 | import numpy as np |
| 6 | 5 | |
| 7 | 6 | from . import _compat as compat |
| 7 | from ._decorator import doc | |
| 8 | 8 | |
| 9 | 9 | |
| 10 | 10 | def _get_sindex_class(): |
| 167 | 167 | """ |
| 168 | 168 | raise NotImplementedError |
| 169 | 169 | |
| 170 | def nearest( | |
| 171 | self, geometry, return_all=True, max_distance=None, return_distance=False | |
| 172 | ): | |
| 173 | """ | |
| 174 | Return the nearest geometry in the tree for each input geometry in | |
| 175 | ``geometry``. | |
| 176 | ||
| 177 | .. note:: | |
| 178 | ``nearest`` currently only works with PyGEOS >= 0.10. | |
| 179 | ||
| 180 | Note that if PyGEOS is not available, geopandas will use rtree | |
| 181 | for the spatial index, where nearest has a different | |
| 182 | function signature to temporarily preserve existing | |
| 183 | functionality. See the documentation of | |
| 184 | :meth:`rtree.index.Index.nearest` for the details on the | |
| 185 | ``rtree``-based implementation. | |
| 186 | ||
| 187 | If multiple tree geometries have the same distance from an input geometry, | |
| 188 | multiple results will be returned for that input geometry by default. | |
| 189 | Specify ``return_all=False`` to only get a single nearest geometry | |
| 190 | (non-deterministic which nearest is returned). | |
| 191 | ||
| 192 | In the context of a spatial join, input geometries are the "left" | |
| 193 | geometries that determine the order of the results, and tree geometries | |
| 194 | are "right" geometries that are joined against the left geometries. | |
| 195 | If ``max_distance`` is not set, this will effectively be a left join | |
| 196 | because every geometry in ``geometry`` will have a nearest geometry in | |
| 197 | the tree. However, if ``max_distance`` is used, this becomes an | |
| 198 | inner join, since some geometries in ``geometry`` may not have a match | |
| 199 | in the tree. | |
| 200 | ||
| 201 | For performance reasons, it is highly recommended that you set | |
| 202 | the ``max_distance`` parameter. | |
| 203 | ||
| 204 | Parameters | |
| 205 | ---------- | |
| 206 | geometry : {shapely.geometry, GeoSeries, GeometryArray, numpy.array of PyGEOS \ | |
| 207 | geometries} | |
| 208 | A single shapely geometry, one of the GeoPandas geometry iterables | |
| 209 | (GeoSeries, GeometryArray), or a numpy array of PyGEOS geometries to query | |
| 210 | against the spatial index. | |
| 211 | return_all : bool, default True | |
| 212 | If there are multiple equidistant or intersecting nearest | |
| 213 | geometries, return all those geometries instead of a single | |
| 214 | nearest geometry. | |
| 215 | max_distance : float, optional | |
| 216 | Maximum distance within which to query for nearest items in tree. | |
| 217 | Must be greater than 0. By default None, indicating no distance limit. | |
| 218 | return_distance : bool, optional | |
| 219 | If True, will return distances in addition to indexes. By default False | |
| 220 | ||
| 221 | Returns | |
| 222 | ------- | |
| 223 | Indices or tuple of (indices, distances) | |
| 224 | Indices is an ndarray of shape (2,n) and distances (if present) an | |
| 225 | ndarray of shape (n). | |
| 226 | The first subarray of indices contains input geometry indices. | |
| 227 | The second subarray of indices contains tree geometry indices. | |
| 228 | ||
| 229 | Examples | |
| 230 | -------- | |
| 231 | >>> from shapely.geometry import Point, box | |
| 232 | >>> s = geopandas.GeoSeries(geopandas.points_from_xy(range(10), range(10))) | |
| 233 | >>> s.head() | |
| 234 | 0 POINT (0.00000 0.00000) | |
| 235 | 1 POINT (1.00000 1.00000) | |
| 236 | 2 POINT (2.00000 2.00000) | |
| 237 | 3 POINT (3.00000 3.00000) | |
| 238 | 4 POINT (4.00000 4.00000) | |
| 239 | dtype: geometry | |
| 240 | ||
| 241 | >>> s.sindex.nearest(Point(1, 1)) | |
| 242 | array([[0], | |
| 243 | [1]]) | |
| 244 | ||
| 245 | >>> s.sindex.nearest([box(4.9, 4.9, 5.1, 5.1)]) | |
| 246 | array([[0], | |
| 247 | [5]]) | |
| 248 | ||
| 249 | >>> s2 = geopandas.GeoSeries(geopandas.points_from_xy([7.6, 10], [7.6, 10])) | |
| 250 | >>> s2 | |
| 251 | 0 POINT (7.60000 7.60000) | |
| 252 | 1 POINT (10.00000 10.00000) | |
| 253 | dtype: geometry | |
| 254 | ||
| 255 | >>> s.sindex.nearest(s2) | |
| 256 | array([[0, 1], | |
| 257 | [8, 9]]) | |
| 258 | """ | |
| 259 | raise NotImplementedError | |
| 260 | ||
| 170 | 261 | def intersection(self, coordinates): |
| 171 | 262 | """Compatibility wrapper for rtree.index.Index.intersection, |
| 172 | use ``query`` intead. | |
| 263 | use ``query`` instead. | |
| 173 | 264 | |
| 174 | 265 | Parameters |
| 175 | 266 | ---------- |
| 264 | 355 | raise NotImplementedError |
| 265 | 356 | |
| 266 | 357 | |
| 267 | def doc(docstring): | |
| 268 | """ | |
| 269 | A decorator take docstring from passed object and it to decorated one. | |
| 270 | """ | |
| 271 | ||
| 272 | def decorator(decorated): | |
| 273 | decorated.__doc__ = dedent(docstring.__doc__ or "") | |
| 274 | return decorated | |
| 275 | ||
| 276 | return decorator | |
| 277 | ||
| 278 | ||
| 279 | 358 | if compat.HAS_RTREE: |
| 280 | 359 | |
| 281 | 360 | import rtree.index # noqa |
| 299 | 378 | def intersection(self, coordinates, *args, **kwargs): |
| 300 | 379 | return super().intersection(coordinates, *args, **kwargs) |
| 301 | 380 | |
| 381 | @doc(BaseSpatialIndex.nearest) | |
| 382 | def nearest(self, *args, **kwargs): | |
| 383 | return super().nearest(*args, **kwargs) | |
| 384 | ||
| 385 | @property | |
| 302 | 386 | @doc(BaseSpatialIndex.size) |
| 303 | @property | |
| 304 | 387 | def size(self): |
| 305 | 388 | return len(self.leaves()[0][1]) |
| 306 | 389 | |
| 390 | @property | |
| 307 | 391 | @doc(BaseSpatialIndex.is_empty) |
| 308 | @property | |
| 309 | 392 | def is_empty(self): |
| 310 | 393 | if len(self.leaves()) > 1: |
| 311 | 394 | return False |
| 342 | 425 | [None] * self.geometries.size, dtype=object |
| 343 | 426 | ) |
| 344 | 427 | |
| 428 | @property | |
| 345 | 429 | @doc(BaseSpatialIndex.valid_query_predicates) |
| 346 | @property | |
| 347 | 430 | def valid_query_predicates(self): |
| 348 | 431 | return { |
| 349 | 432 | None, |
| 449 | 532 | input_geometry_index.extend([i] * len(res)) |
| 450 | 533 | return np.vstack([input_geometry_index, tree_index]) |
| 451 | 534 | |
| 535 | def nearest(self, coordinates, num_results=1, objects=False): | |
| 536 | """ | |
| 537 | Returns the nearest object or objects to the given coordinates. | |
| 538 | ||
| 539 | Requires rtree, and passes parameters directly to | |
| 540 | :meth:`rtree.index.Index.nearest`. | |
| 541 | ||
| 542 | This behaviour is deprecated and will be updated to be consistent | |
| 543 | with the pygeos PyGEOSSTRTreeIndex in a future release. | |
| 544 | ||
| 545 | If longer-term compatibility is required, use | |
| 546 | :meth:`rtree.index.Index.nearest` directly instead. | |
| 547 | ||
| 548 | Examples | |
| 549 | -------- | |
| 550 | >>> s = geopandas.GeoSeries(geopandas.points_from_xy(range(3), range(3))) | |
| 551 | >>> s | |
| 552 | 0 POINT (0.00000 0.00000) | |
| 553 | 1 POINT (1.00000 1.00000) | |
| 554 | 2 POINT (2.00000 2.00000) | |
| 555 | dtype: geometry | |
| 556 | ||
| 557 | >>> list(s.sindex.nearest((0, 0))) # doctest: +SKIP | |
| 558 | [0] | |
| 559 | ||
| 560 | >>> list(s.sindex.nearest((0.5, 0.5))) # doctest: +SKIP | |
| 561 | [0, 1] | |
| 562 | ||
| 563 | >>> list(s.sindex.nearest((3, 3), num_results=2)) # doctest: +SKIP | |
| 564 | [2, 1] | |
| 565 | ||
| 566 | >>> list(super(type(s.sindex), s.sindex).nearest((0, 0), | |
| 567 | ... num_results=2)) # doctest: +SKIP | |
| 568 | [0, 1] | |
| 569 | ||
| 570 | Parameters | |
| 571 | ---------- | |
| 572 | coordinates : sequence or array | |
| 573 | This may be an object that satisfies the numpy array protocol, | |
| 574 | providing the index’s dimension * 2 coordinate pairs | |
| 575 | representing the mink and maxk coordinates in each dimension | |
| 576 | defining the bounds of the query window. | |
| 577 | num_results : integer | |
| 578 | The number of results to return nearest to the given | |
| 579 | coordinates. If two index entries are equidistant, both are | |
| 580 | returned. This property means that num_results may return more | |
| 581 | items than specified | |
| 582 | objects : True / False / ‘raw’ | |
| 583 | If True, the nearest method will return index objects that were | |
| 584 | pickled when they were stored with each index entry, as well as | |
| 585 | the id and bounds of the index entries. If ‘raw’, it will | |
| 586 | return the object as entered into the database without the | |
| 587 | rtree.index.Item wrapper. | |
| 588 | """ | |
| 589 | warnings.warn( | |
| 590 | "sindex.nearest using the rtree backend was not previously documented " | |
| 591 | "and this behavior is deprecated in favor of matching the function " | |
| 592 | "signature provided by the pygeos backend (see " | |
| 593 | "PyGEOSSTRTreeIndex.nearest for details). This behavior will be " | |
| 594 | "updated in a future release.", | |
| 595 | FutureWarning, | |
| 596 | ) | |
| 597 | return super().nearest( | |
| 598 | coordinates, num_results=num_results, objects=objects | |
| 599 | ) | |
| 600 | ||
| 452 | 601 | @doc(BaseSpatialIndex.intersection) |
| 453 | 602 | def intersection(self, coordinates): |
| 454 | 603 | return super().intersection(coordinates, objects=False) |
| 455 | 604 | |
| 605 | @property | |
| 456 | 606 | @doc(BaseSpatialIndex.size) |
| 457 | @property | |
| 458 | 607 | def size(self): |
| 459 | 608 | if hasattr(self, "_size"): |
| 460 | 609 | size = self._size |
| 466 | 615 | self._size = size |
| 467 | 616 | return size |
| 468 | 617 | |
| 618 | @property | |
| 469 | 619 | @doc(BaseSpatialIndex.is_empty) |
| 470 | @property | |
| 471 | 620 | def is_empty(self): |
| 472 | 621 | return self.geometries.size == 0 or self.size == 0 |
| 473 | 622 | |
| 480 | 629 | from . import geoseries # noqa |
| 481 | 630 | from . import array # noqa |
| 482 | 631 | import pygeos # noqa |
| 632 | ||
| 633 | _PYGEOS_PREDICATES = {p.name for p in pygeos.strtree.BinaryPredicate} | set([None]) | |
| 483 | 634 | |
| 484 | 635 | class PyGEOSSTRTreeIndex(pygeos.STRtree): |
| 485 | 636 | """A simple wrapper around pygeos's STRTree. |
| 498 | 649 | # https://github.com/pygeos/pygeos/issues/147 |
| 499 | 650 | non_empty = geometry.copy() |
| 500 | 651 | non_empty[pygeos.is_empty(non_empty)] = None |
| 501 | # set empty geometries to None to mantain indexing | |
| 652 | # set empty geometries to None to maintain indexing | |
| 502 | 653 | super().__init__(non_empty) |
| 503 | 654 | # store geometries, including empty geometries for user access |
| 504 | 655 | self.geometries = geometry.copy() |
| 520 | 671 | {'contains', 'crosses', 'covered_by', None, 'intersects', 'within', \ |
| 521 | 672 | 'touches', 'overlaps', 'contains_properly', 'covers'} |
| 522 | 673 | """ |
| 523 | return pygeos.strtree.VALID_PREDICATES | set([None]) | |
| 674 | return _PYGEOS_PREDICATES | |
| 524 | 675 | |
| 525 | 676 | @doc(BaseSpatialIndex.query) |
| 526 | 677 | def query(self, geometry, predicate=None, sort=False): |
| 542 | 693 | |
| 543 | 694 | return matches |
| 544 | 695 | |
| 696 | @staticmethod | |
| 697 | def _as_geometry_array(geometry): | |
| 698 | """Convert geometry into a numpy array of PyGEOS geometries. | |
| 699 | ||
| 700 | Parameters | |
| 701 | ---------- | |
| 702 | geometry | |
| 703 | An array-like of PyGEOS geometries, a GeoPandas GeoSeries/GeometryArray, | |
| 704 | shapely.geometry or list of shapely geometries. | |
| 705 | ||
| 706 | Returns | |
| 707 | ------- | |
| 708 | np.ndarray | |
| 709 | A numpy array of pygeos geometries. | |
| 710 | """ | |
| 711 | if isinstance(geometry, np.ndarray): | |
| 712 | return geometry | |
| 713 | elif isinstance(geometry, geoseries.GeoSeries): | |
| 714 | return geometry.values.data | |
| 715 | elif isinstance(geometry, array.GeometryArray): | |
| 716 | return geometry.data | |
| 717 | elif isinstance(geometry, BaseGeometry): | |
| 718 | return array._shapely_to_geom(geometry) | |
| 719 | elif isinstance(geometry, list): | |
| 720 | return np.asarray( | |
| 721 | [ | |
| 722 | array._shapely_to_geom(el) | |
| 723 | if isinstance(el, BaseGeometry) | |
| 724 | else el | |
| 725 | for el in geometry | |
| 726 | ] | |
| 727 | ) | |
| 728 | else: | |
| 729 | return np.asarray(geometry) | |
| 730 | ||
| 545 | 731 | @doc(BaseSpatialIndex.query_bulk) |
| 546 | 732 | def query_bulk(self, geometry, predicate=None, sort=False): |
| 547 | 733 | if predicate not in self.valid_query_predicates: |
| 550 | 736 | predicate, self.valid_query_predicates |
| 551 | 737 | ) |
| 552 | 738 | ) |
| 553 | if isinstance(geometry, geoseries.GeoSeries): | |
| 554 | geometry = geometry.values.data | |
| 555 | elif isinstance(geometry, array.GeometryArray): | |
| 556 | geometry = geometry.data | |
| 557 | elif not isinstance(geometry, np.ndarray): | |
| 558 | geometry = np.asarray(geometry) | |
| 739 | ||
| 740 | geometry = self._as_geometry_array(geometry) | |
| 559 | 741 | |
| 560 | 742 | res = super().query_bulk(geometry, predicate) |
| 561 | 743 | |
| 566 | 748 | return np.vstack((geo_res[indexing], tree_res[indexing])) |
| 567 | 749 | |
| 568 | 750 | return res |
| 751 | ||
| 752 | @doc(BaseSpatialIndex.nearest) | |
| 753 | def nearest( | |
| 754 | self, geometry, return_all=True, max_distance=None, return_distance=False | |
| 755 | ): | |
| 756 | if not compat.PYGEOS_GE_010: | |
| 757 | raise NotImplementedError("sindex.nearest requires pygeos >= 0.10") | |
| 758 | ||
| 759 | geometry = self._as_geometry_array(geometry) | |
| 760 | ||
| 761 | if not return_all and max_distance is None and not return_distance: | |
| 762 | return super().nearest(geometry) | |
| 763 | ||
| 764 | result = super().nearest_all( | |
| 765 | geometry, max_distance=max_distance, return_distance=return_distance | |
| 766 | ) | |
| 767 | if return_distance: | |
| 768 | indices, distances = result | |
| 769 | else: | |
| 770 | indices = result | |
| 771 | ||
| 772 | if not return_all: | |
| 773 | # first subarray of geometry indices is sorted, so we can use this | |
| 774 | # trick to get the first of each index value | |
| 775 | mask = np.diff(indices[0, :]).astype("bool") | |
| 776 | # always select the first element | |
| 777 | mask = np.insert(mask, 0, True) | |
| 778 | ||
| 779 | indices = indices[:, mask] | |
| 780 | if return_distance: | |
| 781 | distances = distances[mask] | |
| 782 | ||
| 783 | if return_distance: | |
| 784 | return indices, distances | |
| 785 | else: | |
| 786 | return indices | |
| 569 | 787 | |
| 570 | 788 | @doc(BaseSpatialIndex.intersection) |
| 571 | 789 | def intersection(self, coordinates): |
| 597 | 815 | |
| 598 | 816 | return indexes |
| 599 | 817 | |
| 818 | @property | |
| 600 | 819 | @doc(BaseSpatialIndex.size) |
| 601 | @property | |
| 602 | 820 | def size(self): |
| 603 | 821 | return len(self) |
| 604 | 822 | |
| 823 | @property | |
| 605 | 824 | @doc(BaseSpatialIndex.is_empty) |
| 606 | @property | |
| 607 | 825 | def is_empty(self): |
| 608 | 826 | return len(self) == 0 |
| 11 | 11 | |
| 12 | 12 | def _isna(this): |
| 13 | 13 | """isna version that works for both scalars and (Geo)Series""" |
| 14 | if hasattr(this, "isna"): | |
| 15 | return this.isna() | |
| 16 | elif hasattr(this, "isnull"): | |
| 17 | return this.isnull() | |
| 18 | else: | |
| 19 | return pd.isnull(this) | |
| 20 | ||
| 21 | ||
| 22 | def geom_equals(this, that): | |
| 14 | with warnings.catch_warnings(): | |
| 15 | # GeoSeries.isna will raise a warning about no longer returning True | |
| 16 | # for empty geometries. This helper is used below always in combination | |
| 17 | # with an is_empty check to preserve behaviour, and thus we ignore the | |
| 18 | # warning here to avoid it bubbling up to the user | |
| 19 | warnings.filterwarnings( | |
| 20 | "ignore", r"GeoSeries.isna\(\) previously returned", UserWarning | |
| 21 | ) | |
| 22 | if hasattr(this, "isna"): | |
| 23 | return this.isna() | |
| 24 | elif hasattr(this, "isnull"): | |
| 25 | return this.isnull() | |
| 26 | else: | |
| 27 | return pd.isnull(this) | |
| 28 | ||
| 29 | ||
| 30 | def _geom_equals_mask(this, that): | |
| 23 | 31 | """ |
| 24 | 32 | Test for geometric equality. Empty or missing geometries are considered |
| 25 | 33 | equal. |
| 28 | 36 | ---------- |
| 29 | 37 | this, that : arrays of Geo objects (or anything that has an `is_empty` |
| 30 | 38 | attribute) |
| 39 | ||
| 40 | Returns | |
| 41 | ------- | |
| 42 | Series | |
| 43 | boolean Series, True if geometries in left equal geometries in right | |
| 31 | 44 | """ |
| 32 | 45 | |
| 33 | 46 | return ( |
| 34 | 47 | this.geom_equals(that) |
| 35 | 48 | | (this.is_empty & that.is_empty) |
| 36 | 49 | | (_isna(this) & _isna(that)) |
| 37 | ).all() | |
| 38 | ||
| 39 | ||
| 40 | def geom_almost_equals(this, that): | |
| 50 | ) | |
| 51 | ||
| 52 | ||
| 53 | def geom_equals(this, that): | |
| 54 | """ | |
| 55 | Test for geometric equality. Empty or missing geometries are considered | |
| 56 | equal. | |
| 57 | ||
| 58 | Parameters | |
| 59 | ---------- | |
| 60 | this, that : arrays of Geo objects (or anything that has an `is_empty` | |
| 61 | attribute) | |
| 62 | ||
| 63 | Returns | |
| 64 | ------- | |
| 65 | bool | |
| 66 | True if all geometries in left equal geometries in right | |
| 67 | """ | |
| 68 | ||
| 69 | return _geom_equals_mask(this, that).all() | |
| 70 | ||
| 71 | ||
| 72 | def _geom_almost_equals_mask(this, that): | |
| 41 | 73 | """ |
| 42 | 74 | Test for 'almost' geometric equality. Empty or missing geometries |
| 43 | 75 | considered equal. |
| 49 | 81 | ---------- |
| 50 | 82 | this, that : arrays of Geo objects (or anything that has an `is_empty` |
| 51 | 83 | property) |
| 84 | ||
| 85 | Returns | |
| 86 | ------- | |
| 87 | Series | |
| 88 | boolean Series, True if geometries in left almost equal geometries in right | |
| 52 | 89 | """ |
| 53 | 90 | |
| 54 | 91 | return ( |
| 55 | 92 | this.geom_almost_equals(that) |
| 56 | 93 | | (this.is_empty & that.is_empty) |
| 57 | 94 | | (_isna(this) & _isna(that)) |
| 58 | ).all() | |
| 95 | ) | |
| 96 | ||
| 97 | ||
| 98 | def geom_almost_equals(this, that): | |
| 99 | """ | |
| 100 | Test for 'almost' geometric equality. Empty or missing geometries | |
| 101 | considered equal. | |
| 102 | ||
| 103 | This method allows small difference in the coordinates, but this | |
| 104 | requires coordinates be in the same order for all components of a geometry. | |
| 105 | ||
| 106 | Parameters | |
| 107 | ---------- | |
| 108 | this, that : arrays of Geo objects (or anything that has an `is_empty` | |
| 109 | property) | |
| 110 | ||
| 111 | Returns | |
| 112 | ------- | |
| 113 | bool | |
| 114 | True if all geometries in left almost equal geometries in right | |
| 115 | """ | |
| 116 | ||
| 117 | return _geom_almost_equals_mask(this, that).all() | |
| 59 | 118 | |
| 60 | 119 | |
| 61 | 120 | def assert_geoseries_equal( |
| 156 | 215 | ) |
| 157 | 216 | if check_less_precise: |
| 158 | 217 | precise = "almost " |
| 159 | if not geom_almost_equals(left, right): | |
| 160 | unequal_left_geoms = left[~left.geom_almost_equals(right)] | |
| 161 | unequal_right_geoms = right[~left.geom_almost_equals(right)] | |
| 162 | raise AssertionError( | |
| 163 | assert_error_message.format( | |
| 164 | len(unequal_left_geoms), | |
| 165 | len(left), | |
| 166 | unequal_left_geoms.index.to_list(), | |
| 167 | precise, | |
| 168 | _truncated_string(unequal_left_geoms.iloc[0]), | |
| 169 | _truncated_string(unequal_right_geoms.iloc[0]), | |
| 170 | ) | |
| 218 | equal = _geom_almost_equals_mask(left, right) | |
| 219 | else: | |
| 220 | precise = "" | |
| 221 | equal = _geom_equals_mask(left, right) | |
| 222 | ||
| 223 | if not equal.all(): | |
| 224 | unequal_left_geoms = left[~equal] | |
| 225 | unequal_right_geoms = right[~equal] | |
| 226 | raise AssertionError( | |
| 227 | assert_error_message.format( | |
| 228 | len(unequal_left_geoms), | |
| 229 | len(left), | |
| 230 | unequal_left_geoms.index.to_list(), | |
| 231 | precise, | |
| 232 | _truncated_string(unequal_left_geoms.iloc[0]), | |
| 233 | _truncated_string(unequal_right_geoms.iloc[0]), | |
| 171 | 234 | ) |
| 172 | else: | |
| 173 | precise = "" | |
| 174 | if not geom_equals(left, right): | |
| 175 | unequal_left_geoms = left[~left.geom_almost_equals(right)] | |
| 176 | unequal_right_geoms = right[~left.geom_almost_equals(right)] | |
| 177 | raise AssertionError( | |
| 178 | assert_error_message.format( | |
| 179 | len(unequal_left_geoms), | |
| 180 | len(left), | |
| 181 | unequal_left_geoms.index.to_list(), | |
| 182 | precise, | |
| 183 | _truncated_string(unequal_left_geoms.iloc[0]), | |
| 184 | _truncated_string(unequal_right_geoms.iloc[0]), | |
| 185 | ) | |
| 186 | ) | |
| 235 | ) | |
| 187 | 236 | |
| 188 | 237 | |
| 189 | 238 | def assert_geodataframe_equal( |
| 138 | 138 | assert all(v.equals(t) for v, t in zip(res, points_no_missing)) |
| 139 | 139 | |
| 140 | 140 | # missing values |
| 141 | # TODO(pygeos) does not support empty strings | |
| 142 | if compat.USE_PYGEOS: | |
| 143 | L_wkb.extend([None]) | |
| 144 | else: | |
| 145 | L_wkb.extend([b"", None]) | |
| 146 | res = from_wkb(L_wkb) | |
| 147 | assert res[-1] is None | |
| 141 | # TODO(pygeos) does not support empty strings, np.nan, or pd.NA | |
| 142 | missing_values = [None] | |
| 148 | 143 | if not compat.USE_PYGEOS: |
| 149 | assert res[-2] is None | |
| 144 | missing_values.extend([b"", np.nan]) | |
| 145 | ||
| 146 | if compat.PANDAS_GE_10: | |
| 147 | missing_values.append(pd.NA) | |
| 148 | ||
| 149 | res = from_wkb(missing_values) | |
| 150 | np.testing.assert_array_equal(res, np.full(len(missing_values), None)) | |
| 150 | 151 | |
| 151 | 152 | # single MultiPolygon |
| 152 | 153 | multi_poly = shapely.geometry.MultiPolygon( |
| 154 | 155 | ) |
| 155 | 156 | res = from_wkb([multi_poly.wkb]) |
| 156 | 157 | assert res[0] == multi_poly |
| 158 | ||
| 159 | ||
| 160 | def test_from_wkb_hex(): | |
| 161 | geometry_hex = ["0101000000CDCCCCCCCCCC1440CDCCCCCCCC0C4A40"] | |
| 162 | res = from_wkb(geometry_hex) | |
| 163 | assert isinstance(res, GeometryArray) | |
| 164 | ||
| 165 | # array | |
| 166 | res = from_wkb(np.array(geometry_hex, dtype=object)) | |
| 167 | assert isinstance(res, GeometryArray) | |
| 157 | 168 | |
| 158 | 169 | |
| 159 | 170 | def test_to_wkb(): |
| 201 | 212 | assert all(v.almost_equals(t) for v, t in zip(res, points_no_missing)) |
| 202 | 213 | |
| 203 | 214 | # missing values |
| 204 | # TODO(pygeos) does not support empty strings | |
| 205 | if compat.USE_PYGEOS: | |
| 206 | L_wkt.extend([None]) | |
| 207 | else: | |
| 208 | L_wkt.extend([f(""), None]) | |
| 209 | res = from_wkt(L_wkt) | |
| 210 | assert res[-1] is None | |
| 215 | # TODO(pygeos) does not support empty strings, np.nan, or pd.NA | |
| 216 | missing_values = [None] | |
| 211 | 217 | if not compat.USE_PYGEOS: |
| 212 | assert res[-2] is None | |
| 218 | missing_values.extend([f(""), np.nan]) | |
| 219 | ||
| 220 | if compat.PANDAS_GE_10: | |
| 221 | missing_values.append(pd.NA) | |
| 222 | ||
| 223 | res = from_wkb(missing_values) | |
| 224 | np.testing.assert_array_equal(res, np.full(len(missing_values), None)) | |
| 213 | 225 | |
| 214 | 226 | # single MultiPolygon |
| 215 | 227 | multi_poly = shapely.geometry.MultiPolygon( |
| 444 | 456 | |
| 445 | 457 | |
| 446 | 458 | @pytest.mark.parametrize( |
| 447 | "attr", ["is_closed", "is_valid", "is_empty", "is_simple", "has_z", "is_ring"] | |
| 459 | "attr", | |
| 460 | [ | |
| 461 | "is_closed", | |
| 462 | "is_valid", | |
| 463 | "is_empty", | |
| 464 | "is_simple", | |
| 465 | "has_z", | |
| 466 | # for is_ring we raise a warning about the value for Polygon changing | |
| 467 | pytest.param( | |
| 468 | "is_ring", marks=pytest.mark.filterwarnings("ignore:is_ring:FutureWarning") | |
| 469 | ), | |
| 470 | ], | |
| 448 | 471 | ) |
| 449 | 472 | def test_unary_predicates(attr): |
| 450 | 473 | na_value = False |
| 481 | 504 | assert result.tolist() == expected |
| 482 | 505 | |
| 483 | 506 | |
| 507 | # for is_ring we raise a warning about the value for Polygon changing | |
| 508 | @pytest.mark.filterwarnings("ignore:is_ring:FutureWarning") | |
| 484 | 509 | def test_is_ring(): |
| 485 | 510 | g = [ |
| 486 | 511 | shapely.geometry.LinearRing([(0, 0), (1, 1), (1, -1)]), |
| 922 | 947 | "EPSG:32618" |
| 923 | 948 | ) |
| 924 | 949 | |
| 950 | @pytest.mark.skipif(not compat.PYPROJ_GE_31, reason="requires pyproj 3.1 or higher") | |
| 951 | def test_estimate_utm_crs__antimeridian(self): | |
| 952 | antimeridian = from_shapely( | |
| 953 | [ | |
| 954 | shapely.geometry.Point(1722483.900174921, 5228058.6143420935), | |
| 955 | shapely.geometry.Point(4624385.494808555, 8692574.544944234), | |
| 956 | ], | |
| 957 | crs="EPSG:3851", | |
| 958 | ) | |
| 959 | assert antimeridian.estimate_utm_crs() == CRS("EPSG:32760") | |
| 960 | ||
| 925 | 961 | @pytest.mark.skipif(compat.PYPROJ_LT_3, reason="requires pyproj 3 or higher") |
| 926 | 962 | def test_estimate_utm_crs__out_of_bounds(self): |
| 927 | 963 | with pytest.raises(RuntimeError, match="Unable to determine UTM CRS"): |
| 219 | 219 | assert df.geometry.values.crs == self.osgb |
| 220 | 220 | |
| 221 | 221 | # different passed CRS than array CRS is ignored |
| 222 | with pytest.warns(FutureWarning): | |
| 222 | with pytest.warns(FutureWarning, match="CRS mismatch"): | |
| 223 | 223 | df = GeoDataFrame(geometry=s, crs=4326) |
| 224 | 224 | assert df.crs == self.osgb |
| 225 | 225 | assert df.geometry.crs == self.osgb |
| 226 | 226 | assert df.geometry.values.crs == self.osgb |
| 227 | with pytest.warns(FutureWarning): | |
| 227 | with pytest.warns(FutureWarning, match="CRS mismatch"): | |
| 228 | 228 | GeoDataFrame(geometry=s, crs=4326) |
| 229 | with pytest.warns(FutureWarning): | |
| 229 | with pytest.warns(FutureWarning, match="CRS mismatch"): | |
| 230 | 230 | GeoDataFrame({"data": [1, 2], "geometry": s}, crs=4326) |
| 231 | with pytest.warns(FutureWarning): | |
| 231 | with pytest.warns(FutureWarning, match="CRS mismatch"): | |
| 232 | 232 | GeoDataFrame(df, crs=4326).crs |
| 233 | 233 | |
| 234 | 234 | # manually change CRS |
| 267 | 267 | assert df.geometry.crs == self.wgs |
| 268 | 268 | assert df.geometry.values.crs == self.wgs |
| 269 | 269 | |
| 270 | arr = from_shapely(self.geoms) | |
| 271 | s = GeoSeries(arr, crs=27700) | |
| 270 | 272 | df = GeoDataFrame() |
| 271 | 273 | df = df.set_geometry(s) |
| 272 | 274 | assert df.crs == self.osgb |
| 296 | 298 | df = GeoDataFrame({"geometry": [0, 1]}) |
| 297 | 299 | df.crs = 27700 |
| 298 | 300 | assert df.crs == self.osgb |
| 301 | ||
| 302 | def test_dataframe_setitem(self): | |
| 303 | # new geometry CRS has priority over GDF CRS | |
| 304 | arr = from_shapely(self.geoms) | |
| 305 | s = GeoSeries(arr, crs=27700) | |
| 306 | df = GeoDataFrame() | |
| 307 | df["geometry"] = s | |
| 308 | assert df.crs == self.osgb | |
| 309 | assert df.geometry.crs == self.osgb | |
| 310 | assert df.geometry.values.crs == self.osgb | |
| 311 | ||
| 312 | arr = from_shapely(self.geoms, crs=27700) | |
| 313 | df = GeoDataFrame() | |
| 314 | df["geometry"] = arr | |
| 315 | assert df.crs == self.osgb | |
| 316 | assert df.geometry.crs == self.osgb | |
| 317 | assert df.geometry.values.crs == self.osgb | |
| 318 | ||
| 319 | # test to_crs case (GH1960) | |
| 320 | arr = from_shapely(self.geoms) | |
| 321 | df = GeoDataFrame({"col1": [1, 2], "geometry": arr}, crs=4326) | |
| 322 | df["geometry"] = df["geometry"].to_crs(27700) | |
| 323 | assert df.crs == self.osgb | |
| 324 | assert df.geometry.crs == self.osgb | |
| 325 | assert df.geometry.values.crs == self.osgb | |
| 326 | ||
| 327 | # test changing geometry crs not in the geometry column doesn't change the crs | |
| 328 | arr = from_shapely(self.geoms) | |
| 329 | df = GeoDataFrame( | |
| 330 | {"col1": [1, 2], "geometry": arr, "other_geom": arr}, crs=4326 | |
| 331 | ) | |
| 332 | df["other_geom"] = from_shapely(self.geoms, crs=27700) | |
| 333 | assert df.crs == self.wgs | |
| 334 | assert df.geometry.crs == self.wgs | |
| 335 | assert df["geometry"].crs == self.wgs | |
| 336 | assert df["other_geom"].crs == self.osgb | |
| 299 | 337 | |
| 300 | 338 | @pytest.mark.parametrize( |
| 301 | 339 | "scalar", [None, Point(0, 0), LineString([(0, 0), (1, 1)])] |
| 0 | from textwrap import dedent | |
| 1 | ||
| 2 | from geopandas._decorator import doc | |
| 3 | ||
| 4 | ||
| 5 | @doc(method="cumsum", operation="sum") | |
| 6 | def cumsum(whatever): | |
| 7 | """ | |
| 8 | This is the {method} method. | |
| 9 | ||
| 10 | It computes the cumulative {operation}. | |
| 11 | """ | |
| 12 | ... | |
| 13 | ||
| 14 | ||
| 15 | @doc( | |
| 16 | cumsum, | |
| 17 | dedent( | |
| 18 | """ | |
| 19 | Examples | |
| 20 | -------- | |
| 21 | ||
| 22 | >>> cumavg([1, 2, 3]) | |
| 23 | 2 | |
| 24 | """ | |
| 25 | ), | |
| 26 | method="cumavg", | |
| 27 | operation="average", | |
| 28 | ) | |
| 29 | def cumavg(whatever): | |
| 30 | ... | |
| 31 | ||
| 32 | ||
| 33 | @doc(cumsum, method="cummax", operation="maximum") | |
| 34 | def cummax(whatever): | |
| 35 | ... | |
| 36 | ||
| 37 | ||
| 38 | @doc(cummax, method="cummin", operation="minimum") | |
| 39 | def cummin(whatever): | |
| 40 | ... | |
| 41 | ||
| 42 | ||
| 43 | def test_docstring_formatting(): | |
| 44 | docstr = dedent( | |
| 45 | """ | |
| 46 | This is the cumsum method. | |
| 47 | ||
| 48 | It computes the cumulative sum. | |
| 49 | """ | |
| 50 | ) | |
| 51 | assert cumsum.__doc__ == docstr | |
| 52 | ||
| 53 | ||
| 54 | def test_docstring_appending(): | |
| 55 | docstr = dedent( | |
| 56 | """ | |
| 57 | This is the cumavg method. | |
| 58 | ||
| 59 | It computes the cumulative average. | |
| 60 | ||
| 61 | Examples | |
| 62 | -------- | |
| 63 | ||
| 64 | >>> cumavg([1, 2, 3]) | |
| 65 | 2 | |
| 66 | """ | |
| 67 | ) | |
| 68 | assert cumavg.__doc__ == docstr | |
| 69 | ||
| 70 | ||
| 71 | def test_doc_template_from_func(): | |
| 72 | docstr = dedent( | |
| 73 | """ | |
| 74 | This is the cummax method. | |
| 75 | ||
| 76 | It computes the cumulative maximum. | |
| 77 | """ | |
| 78 | ) | |
| 79 | assert cummax.__doc__ == docstr | |
| 80 | ||
| 81 | ||
| 82 | def test_inherit_doc_template(): | |
| 83 | docstr = dedent( | |
| 84 | """ | |
| 85 | This is the cummin method. | |
| 86 | ||
| 87 | It computes the cumulative minimum. | |
| 88 | """ | |
| 89 | ) | |
| 90 | assert cummin.__doc__ == docstr |
| 200 | 200 | assert_frame_equal(expected_unsorted, gdf.dissolve("a", sort=False)) |
| 201 | 201 | |
| 202 | 202 | |
| 203 | @pytest.mark.skipif( | |
| 204 | not compat.PANDAS_GE_025, | |
| 205 | reason="'observed' param behavior changed in pandas 0.25.0", | |
| 206 | ) | |
| 207 | 203 | def test_dissolve_categorical(): |
| 208 | 204 | gdf = geopandas.GeoDataFrame( |
| 209 | 205 | { |
| 0 | import geopandas as gpd | |
| 1 | import numpy as np | |
| 2 | import pandas as pd | |
| 3 | import pytest | |
| 4 | from distutils.version import LooseVersion | |
| 5 | ||
| 6 | folium = pytest.importorskip("folium") | |
| 7 | branca = pytest.importorskip("branca") | |
| 8 | matplotlib = pytest.importorskip("matplotlib") | |
| 9 | mapclassify = pytest.importorskip("mapclassify") | |
| 10 | ||
| 11 | import matplotlib.cm as cm # noqa | |
| 12 | import matplotlib.colors as colors # noqa | |
| 13 | from branca.colormap import StepColormap # noqa | |
| 14 | ||
| 15 | BRANCA_05 = str(branca.__version__) > LooseVersion("0.4.2") | |
| 16 | ||
| 17 | ||
| 18 | class TestExplore: | |
| 19 | def setup_method(self): | |
| 20 | self.nybb = gpd.read_file(gpd.datasets.get_path("nybb")) | |
| 21 | self.world = gpd.read_file(gpd.datasets.get_path("naturalearth_lowres")) | |
| 22 | self.cities = gpd.read_file(gpd.datasets.get_path("naturalearth_cities")) | |
| 23 | self.world["range"] = range(len(self.world)) | |
| 24 | self.missing = self.world.copy() | |
| 25 | np.random.seed(42) | |
| 26 | self.missing.loc[np.random.choice(self.missing.index, 40), "continent"] = np.nan | |
| 27 | self.missing.loc[np.random.choice(self.missing.index, 40), "pop_est"] = np.nan | |
| 28 | ||
| 29 | def _fetch_map_string(self, m): | |
| 30 | out = m._parent.render() | |
| 31 | out_str = "".join(out.split()) | |
| 32 | return out_str | |
| 33 | ||
| 34 | def test_simple_pass(self): | |
| 35 | """Make sure default pass""" | |
| 36 | self.nybb.explore() | |
| 37 | self.world.explore() | |
| 38 | self.cities.explore() | |
| 39 | self.world.geometry.explore() | |
| 40 | ||
| 41 | def test_choropleth_pass(self): | |
| 42 | """Make sure default choropleth pass""" | |
| 43 | self.world.explore(column="pop_est") | |
| 44 | ||
| 45 | def test_map_settings_default(self): | |
| 46 | """Check default map settings""" | |
| 47 | m = self.world.explore() | |
| 48 | assert m.location == [ | |
| 49 | pytest.approx(-3.1774349999999956, rel=1e-6), | |
| 50 | pytest.approx(2.842170943040401e-14, rel=1e-6), | |
| 51 | ] | |
| 52 | assert m.options["zoom"] == 10 | |
| 53 | assert m.options["zoomControl"] is True | |
| 54 | assert m.position == "relative" | |
| 55 | assert m.height == (100.0, "%") | |
| 56 | assert m.width == (100.0, "%") | |
| 57 | assert m.left == (0, "%") | |
| 58 | assert m.top == (0, "%") | |
| 59 | assert m.global_switches.no_touch is False | |
| 60 | assert m.global_switches.disable_3d is False | |
| 61 | assert "openstreetmap" in m.to_dict()["children"].keys() | |
| 62 | ||
| 63 | def test_map_settings_custom(self): | |
| 64 | """Check custom map settins""" | |
| 65 | m = self.nybb.explore( | |
| 66 | zoom_control=False, | |
| 67 | width=200, | |
| 68 | height=200, | |
| 69 | ) | |
| 70 | assert m.location == [ | |
| 71 | pytest.approx(40.70582377450201, rel=1e-6), | |
| 72 | pytest.approx(-73.9778006856748, rel=1e-6), | |
| 73 | ] | |
| 74 | assert m.options["zoom"] == 10 | |
| 75 | assert m.options["zoomControl"] is False | |
| 76 | assert m.height == (200.0, "px") | |
| 77 | assert m.width == (200.0, "px") | |
| 78 | ||
| 79 | # custom XYZ tiles | |
| 80 | m = self.nybb.explore( | |
| 81 | zoom_control=False, | |
| 82 | width=200, | |
| 83 | height=200, | |
| 84 | tiles="https://mt1.google.com/vt/lyrs=m&x={x}&y={y}&z={z}", | |
| 85 | attr="Google", | |
| 86 | ) | |
| 87 | ||
| 88 | out_str = self._fetch_map_string(m) | |
| 89 | s = '"https://mt1.google.com/vt/lyrs=m\\u0026x={x}\\u0026y={y}\\u0026z={z}"' | |
| 90 | assert s in out_str | |
| 91 | assert '"attribution":"Google"' in out_str | |
| 92 | ||
| 93 | m = self.nybb.explore(location=(40, 5)) | |
| 94 | assert m.location == [40, 5] | |
| 95 | assert m.options["zoom"] == 10 | |
| 96 | ||
| 97 | m = self.nybb.explore(zoom_start=8) | |
| 98 | assert m.location == [ | |
| 99 | pytest.approx(40.70582377450201, rel=1e-6), | |
| 100 | pytest.approx(-73.9778006856748, rel=1e-6), | |
| 101 | ] | |
| 102 | assert m.options["zoom"] == 8 | |
| 103 | ||
| 104 | m = self.nybb.explore(location=(40, 5), zoom_start=8) | |
| 105 | assert m.location == [40, 5] | |
| 106 | assert m.options["zoom"] == 8 | |
| 107 | ||
| 108 | def test_simple_color(self): | |
| 109 | """Check color settings""" | |
| 110 | # single named color | |
| 111 | m = self.nybb.explore(color="red") | |
| 112 | out_str = self._fetch_map_string(m) | |
| 113 | assert '"fillColor":"red"' in out_str | |
| 114 | ||
| 115 | # list of colors | |
| 116 | colors = ["#333333", "#367324", "#95824f", "#fcaa00", "#ffcc33"] | |
| 117 | m2 = self.nybb.explore(color=colors) | |
| 118 | out_str = self._fetch_map_string(m2) | |
| 119 | for c in colors: | |
| 120 | assert f'"fillColor":"{c}"' in out_str | |
| 121 | ||
| 122 | # column of colors | |
| 123 | df = self.nybb.copy() | |
| 124 | df["colors"] = colors | |
| 125 | m3 = df.explore(color="colors") | |
| 126 | out_str = self._fetch_map_string(m3) | |
| 127 | for c in colors: | |
| 128 | assert f'"fillColor":"{c}"' in out_str | |
| 129 | ||
| 130 | # line GeoSeries | |
| 131 | m4 = self.nybb.boundary.explore(color="red") | |
| 132 | out_str = self._fetch_map_string(m4) | |
| 133 | assert '"fillColor":"red"' in out_str | |
| 134 | ||
| 135 | def test_choropleth_linear(self): | |
| 136 | """Check choropleth colors""" | |
| 137 | # default cmap | |
| 138 | m = self.nybb.explore(column="Shape_Leng") | |
| 139 | out_str = self._fetch_map_string(m) | |
| 140 | assert 'color":"#440154"' in out_str | |
| 141 | assert 'color":"#fde725"' in out_str | |
| 142 | assert 'color":"#50c46a"' in out_str | |
| 143 | assert 'color":"#481467"' in out_str | |
| 144 | assert 'color":"#3d4e8a"' in out_str | |
| 145 | ||
| 146 | # named cmap | |
| 147 | m = self.nybb.explore(column="Shape_Leng", cmap="PuRd") | |
| 148 | out_str = self._fetch_map_string(m) | |
| 149 | assert 'color":"#f7f4f9"' in out_str | |
| 150 | assert 'color":"#67001f"' in out_str | |
| 151 | assert 'color":"#d31760"' in out_str | |
| 152 | assert 'color":"#f0ecf5"' in out_str | |
| 153 | assert 'color":"#d6bedc"' in out_str | |
| 154 | ||
| 155 | def test_choropleth_mapclassify(self): | |
| 156 | """Mapclassify bins""" | |
| 157 | # quantiles | |
| 158 | m = self.nybb.explore(column="Shape_Leng", scheme="quantiles") | |
| 159 | out_str = self._fetch_map_string(m) | |
| 160 | assert 'color":"#21918c"' in out_str | |
| 161 | assert 'color":"#3b528b"' in out_str | |
| 162 | assert 'color":"#5ec962"' in out_str | |
| 163 | assert 'color":"#fde725"' in out_str | |
| 164 | assert 'color":"#440154"' in out_str | |
| 165 | ||
| 166 | # headtail | |
| 167 | m = self.world.explore(column="pop_est", scheme="headtailbreaks") | |
| 168 | out_str = self._fetch_map_string(m) | |
| 169 | assert '"fillColor":"#3b528b"' in out_str | |
| 170 | assert '"fillColor":"#21918c"' in out_str | |
| 171 | assert '"fillColor":"#5ec962"' in out_str | |
| 172 | assert '"fillColor":"#fde725"' in out_str | |
| 173 | assert '"fillColor":"#440154"' in out_str | |
| 174 | # custom k | |
| 175 | m = self.world.explore(column="pop_est", scheme="naturalbreaks", k=3) | |
| 176 | out_str = self._fetch_map_string(m) | |
| 177 | assert '"fillColor":"#21918c"' in out_str | |
| 178 | assert '"fillColor":"#fde725"' in out_str | |
| 179 | assert '"fillColor":"#440154"' in out_str | |
| 180 | ||
| 181 | def test_categorical(self): | |
| 182 | """Categorical maps""" | |
| 183 | # auto detection | |
| 184 | m = self.world.explore(column="continent") | |
| 185 | out_str = self._fetch_map_string(m) | |
| 186 | assert 'color":"#9467bd","continent":"Europe"' in out_str | |
| 187 | assert 'color":"#c49c94","continent":"NorthAmerica"' in out_str | |
| 188 | assert 'color":"#1f77b4","continent":"Africa"' in out_str | |
| 189 | assert 'color":"#98df8a","continent":"Asia"' in out_str | |
| 190 | assert 'color":"#ff7f0e","continent":"Antarctica"' in out_str | |
| 191 | assert 'color":"#9edae5","continent":"SouthAmerica"' in out_str | |
| 192 | assert 'color":"#7f7f7f","continent":"Oceania"' in out_str | |
| 193 | assert 'color":"#dbdb8d","continent":"Sevenseas(openocean)"' in out_str | |
| 194 | ||
| 195 | # forced categorical | |
| 196 | m = self.nybb.explore(column="BoroCode", categorical=True) | |
| 197 | out_str = self._fetch_map_string(m) | |
| 198 | assert 'color":"#9edae5"' in out_str | |
| 199 | assert 'color":"#c7c7c7"' in out_str | |
| 200 | assert 'color":"#8c564b"' in out_str | |
| 201 | assert 'color":"#1f77b4"' in out_str | |
| 202 | assert 'color":"#98df8a"' in out_str | |
| 203 | ||
| 204 | # pandas.Categorical | |
| 205 | df = self.world.copy() | |
| 206 | df["categorical"] = pd.Categorical(df["name"]) | |
| 207 | m = df.explore(column="categorical") | |
| 208 | out_str = self._fetch_map_string(m) | |
| 209 | for c in np.apply_along_axis(colors.to_hex, 1, cm.tab20(range(20))): | |
| 210 | assert f'"fillColor":"{c}"' in out_str | |
| 211 | ||
| 212 | # custom cmap | |
| 213 | m = self.nybb.explore(column="BoroName", cmap="Set1") | |
| 214 | out_str = self._fetch_map_string(m) | |
| 215 | assert 'color":"#999999"' in out_str | |
| 216 | assert 'color":"#a65628"' in out_str | |
| 217 | assert 'color":"#4daf4a"' in out_str | |
| 218 | assert 'color":"#e41a1c"' in out_str | |
| 219 | assert 'color":"#ff7f00"' in out_str | |
| 220 | ||
| 221 | # custom list of colors | |
| 222 | cmap = ["#333432", "#3b6e8c", "#bc5b4f", "#8fa37e", "#efc758"] | |
| 223 | m = self.nybb.explore(column="BoroName", cmap=cmap) | |
| 224 | out_str = self._fetch_map_string(m) | |
| 225 | for c in cmap: | |
| 226 | assert f'"fillColor":"{c}"' in out_str | |
| 227 | ||
| 228 | # shorter list (to make it repeat) | |
| 229 | cmap = ["#333432", "#3b6e8c"] | |
| 230 | m = self.nybb.explore(column="BoroName", cmap=cmap) | |
| 231 | out_str = self._fetch_map_string(m) | |
| 232 | for c in cmap: | |
| 233 | assert f'"fillColor":"{c}"' in out_str | |
| 234 | ||
| 235 | with pytest.raises(ValueError, match="'cmap' is invalid."): | |
| 236 | self.nybb.explore(column="BoroName", cmap="nonsense") | |
| 237 | ||
| 238 | def test_categories(self): | |
| 239 | m = self.nybb[["BoroName", "geometry"]].explore( | |
| 240 | column="BoroName", | |
| 241 | categories=["Brooklyn", "Staten Island", "Queens", "Bronx", "Manhattan"], | |
| 242 | ) | |
| 243 | out_str = self._fetch_map_string(m) | |
| 244 | assert '"Bronx","__folium_color":"#c7c7c7"' in out_str | |
| 245 | assert '"Manhattan","__folium_color":"#9edae5"' in out_str | |
| 246 | assert '"Brooklyn","__folium_color":"#1f77b4"' in out_str | |
| 247 | assert '"StatenIsland","__folium_color":"#98df8a"' in out_str | |
| 248 | assert '"Queens","__folium_color":"#8c564b"' in out_str | |
| 249 | ||
| 250 | df = self.nybb.copy() | |
| 251 | df["categorical"] = pd.Categorical(df["BoroName"]) | |
| 252 | with pytest.raises(ValueError, match="Cannot specify 'categories'"): | |
| 253 | df.explore("categorical", categories=["Brooklyn", "Staten Island"]) | |
| 254 | ||
| 255 | def test_column_values(self): | |
| 256 | """ | |
| 257 | Check that the dataframe plot method returns same values with an | |
| 258 | input string (column in df), pd.Series, or np.array | |
| 259 | """ | |
| 260 | column_array = np.array(self.world["pop_est"]) | |
| 261 | m1 = self.world.explore(column="pop_est") # column name | |
| 262 | m2 = self.world.explore(column=column_array) # np.array | |
| 263 | m3 = self.world.explore(column=self.world["pop_est"]) # pd.Series | |
| 264 | assert m1.location == m2.location == m3.location | |
| 265 | ||
| 266 | m1_fields = self.world.explore(column=column_array, tooltip=True, popup=True) | |
| 267 | out1_fields_str = self._fetch_map_string(m1_fields) | |
| 268 | assert ( | |
| 269 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 270 | in out1_fields_str | |
| 271 | ) | |
| 272 | assert ( | |
| 273 | 'aliases=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 274 | in out1_fields_str | |
| 275 | ) | |
| 276 | ||
| 277 | m2_fields = self.world.explore( | |
| 278 | column=self.world["pop_est"], tooltip=True, popup=True | |
| 279 | ) | |
| 280 | out2_fields_str = self._fetch_map_string(m2_fields) | |
| 281 | assert ( | |
| 282 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 283 | in out2_fields_str | |
| 284 | ) | |
| 285 | assert ( | |
| 286 | 'aliases=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 287 | in out2_fields_str | |
| 288 | ) | |
| 289 | ||
| 290 | # GeoDataframe and the given list have different number of rows | |
| 291 | with pytest.raises(ValueError, match="different number of rows"): | |
| 292 | self.world.explore(column=np.array([1, 2, 3])) | |
| 293 | ||
| 294 | def test_no_crs(self): | |
| 295 | """Naive geometry get no tiles""" | |
| 296 | df = self.world.copy() | |
| 297 | df.crs = None | |
| 298 | m = df.explore() | |
| 299 | assert "openstreetmap" not in m.to_dict()["children"].keys() | |
| 300 | ||
| 301 | def test_style_kwds(self): | |
| 302 | """Style keywords""" | |
| 303 | m = self.world.explore( | |
| 304 | style_kwds=dict(fillOpacity=0.1, weight=0.5, fillColor="orange") | |
| 305 | ) | |
| 306 | out_str = self._fetch_map_string(m) | |
| 307 | assert '"fillColor":"orange","fillOpacity":0.1,"weight":0.5' in out_str | |
| 308 | m = self.world.explore(column="pop_est", style_kwds=dict(color="black")) | |
| 309 | assert '"color":"black"' in self._fetch_map_string(m) | |
| 310 | ||
| 311 | def test_tooltip(self): | |
| 312 | """Test tooltip""" | |
| 313 | # default with no tooltip or popup | |
| 314 | m = self.world.explore() | |
| 315 | assert "GeoJsonTooltip" in str(m.to_dict()) | |
| 316 | assert "GeoJsonPopup" not in str(m.to_dict()) | |
| 317 | ||
| 318 | # True | |
| 319 | m = self.world.explore(tooltip=True, popup=True) | |
| 320 | assert "GeoJsonTooltip" in str(m.to_dict()) | |
| 321 | assert "GeoJsonPopup" in str(m.to_dict()) | |
| 322 | out_str = self._fetch_map_string(m) | |
| 323 | assert ( | |
| 324 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 325 | in out_str | |
| 326 | ) | |
| 327 | assert ( | |
| 328 | 'aliases=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 329 | in out_str | |
| 330 | ) | |
| 331 | ||
| 332 | # True choropleth | |
| 333 | m = self.world.explore(column="pop_est", tooltip=True, popup=True) | |
| 334 | assert "GeoJsonTooltip" in str(m.to_dict()) | |
| 335 | assert "GeoJsonPopup" in str(m.to_dict()) | |
| 336 | out_str = self._fetch_map_string(m) | |
| 337 | assert ( | |
| 338 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 339 | in out_str | |
| 340 | ) | |
| 341 | assert ( | |
| 342 | 'aliases=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 343 | in out_str | |
| 344 | ) | |
| 345 | ||
| 346 | # single column | |
| 347 | m = self.world.explore(tooltip="pop_est", popup="iso_a3") | |
| 348 | out_str = self._fetch_map_string(m) | |
| 349 | assert 'fields=["pop_est"]' in out_str | |
| 350 | assert 'aliases=["pop_est"]' in out_str | |
| 351 | assert 'fields=["iso_a3"]' in out_str | |
| 352 | assert 'aliases=["iso_a3"]' in out_str | |
| 353 | ||
| 354 | # list | |
| 355 | m = self.world.explore( | |
| 356 | tooltip=["pop_est", "continent"], popup=["iso_a3", "gdp_md_est"] | |
| 357 | ) | |
| 358 | out_str = self._fetch_map_string(m) | |
| 359 | assert 'fields=["pop_est","continent"]' in out_str | |
| 360 | assert 'aliases=["pop_est","continent"]' in out_str | |
| 361 | assert 'fields=["iso_a3","gdp_md_est"' in out_str | |
| 362 | assert 'aliases=["iso_a3","gdp_md_est"]' in out_str | |
| 363 | ||
| 364 | # number | |
| 365 | m = self.world.explore(tooltip=2, popup=2) | |
| 366 | out_str = self._fetch_map_string(m) | |
| 367 | assert 'fields=["pop_est","continent"]' in out_str | |
| 368 | assert 'aliases=["pop_est","continent"]' in out_str | |
| 369 | ||
| 370 | # keywords tooltip | |
| 371 | m = self.world.explore( | |
| 372 | tooltip=True, | |
| 373 | popup=False, | |
| 374 | tooltip_kwds=dict(aliases=[0, 1, 2, 3, 4, 5], sticky=False), | |
| 375 | ) | |
| 376 | out_str = self._fetch_map_string(m) | |
| 377 | assert ( | |
| 378 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 379 | in out_str | |
| 380 | ) | |
| 381 | assert "aliases=[0,1,2,3,4,5]" in out_str | |
| 382 | assert '"sticky":false' in out_str | |
| 383 | ||
| 384 | # keywords popup | |
| 385 | m = self.world.explore( | |
| 386 | tooltip=False, | |
| 387 | popup=True, | |
| 388 | popup_kwds=dict(aliases=[0, 1, 2, 3, 4, 5]), | |
| 389 | ) | |
| 390 | out_str = self._fetch_map_string(m) | |
| 391 | assert ( | |
| 392 | 'fields=["pop_est","continent","name","iso_a3","gdp_md_est","range"]' | |
| 393 | in out_str | |
| 394 | ) | |
| 395 | assert "aliases=[0,1,2,3,4,5]" in out_str | |
| 396 | assert "<th>${aliases[i]" in out_str | |
| 397 | ||
| 398 | # no labels | |
| 399 | m = self.world.explore( | |
| 400 | tooltip=True, | |
| 401 | popup=True, | |
| 402 | tooltip_kwds=dict(labels=False), | |
| 403 | popup_kwds=dict(labels=False), | |
| 404 | ) | |
| 405 | out_str = self._fetch_map_string(m) | |
| 406 | assert "<th>${aliases[i]" not in out_str | |
| 407 | ||
| 408 | # named index | |
| 409 | gdf = self.nybb.set_index("BoroName") | |
| 410 | m = gdf.explore() | |
| 411 | out_str = self._fetch_map_string(m) | |
| 412 | assert "BoroName" in out_str | |
| 413 | ||
| 414 | def test_default_markers(self): | |
| 415 | # check overriden default for points | |
| 416 | m = self.cities.explore() | |
| 417 | strings = ['"radius":2', '"fill":true', "CircleMarker(latlng,opts)"] | |
| 418 | out_str = self._fetch_map_string(m) | |
| 419 | for s in strings: | |
| 420 | assert s in out_str | |
| 421 | ||
| 422 | m = self.cities.explore(marker_kwds=dict(radius=5, fill=False)) | |
| 423 | strings = ['"radius":5', '"fill":false', "CircleMarker(latlng,opts)"] | |
| 424 | out_str = self._fetch_map_string(m) | |
| 425 | for s in strings: | |
| 426 | assert s in out_str | |
| 427 | ||
| 428 | def test_custom_markers(self): | |
| 429 | # Markers | |
| 430 | m = self.cities.explore( | |
| 431 | marker_type="marker", | |
| 432 | marker_kwds={"icon": folium.Icon(icon="star")}, | |
| 433 | ) | |
| 434 | assert ""","icon":"star",""" in self._fetch_map_string(m) | |
| 435 | ||
| 436 | # Circle Markers | |
| 437 | m = self.cities.explore(marker_type="circle", marker_kwds={"fill_color": "red"}) | |
| 438 | assert ""","fillColor":"red",""" in self._fetch_map_string(m) | |
| 439 | ||
| 440 | # Folium Markers | |
| 441 | m = self.cities.explore( | |
| 442 | marker_type=folium.Circle( | |
| 443 | radius=4, fill_color="orange", fill_opacity=0.4, color="black", weight=1 | |
| 444 | ), | |
| 445 | ) | |
| 446 | assert ""","color":"black",""" in self._fetch_map_string(m) | |
| 447 | ||
| 448 | # Circle | |
| 449 | m = self.cities.explore(marker_type="circle_marker", marker_kwds={"radius": 10}) | |
| 450 | assert ""","radius":10,""" in self._fetch_map_string(m) | |
| 451 | ||
| 452 | # Unsupported Markers | |
| 453 | with pytest.raises( | |
| 454 | ValueError, | |
| 455 | match="Only 'marker', 'circle', and 'circle_marker' are supported", | |
| 456 | ): | |
| 457 | self.cities.explore(marker_type="dummy") | |
| 458 | ||
| 459 | def test_vmin_vmax(self): | |
| 460 | df = self.world.copy() | |
| 461 | df["range"] = range(len(df)) | |
| 462 | m = df.explore("range", vmin=-100, vmax=1000) | |
| 463 | out_str = self._fetch_map_string(m) | |
| 464 | assert 'case"176":return{"color":"#3b528b","fillColor":"#3b528b"' in out_str | |
| 465 | assert 'case"119":return{"color":"#414287","fillColor":"#414287"' in out_str | |
| 466 | assert 'case"3":return{"color":"#482173","fillColor":"#482173"' in out_str | |
| 467 | ||
| 468 | def test_missing_vals(self): | |
| 469 | m = self.missing.explore("continent") | |
| 470 | assert '"fillColor":null' in self._fetch_map_string(m) | |
| 471 | ||
| 472 | m = self.missing.explore("pop_est") | |
| 473 | assert '"fillColor":null' in self._fetch_map_string(m) | |
| 474 | ||
| 475 | m = self.missing.explore("pop_est", missing_kwds=dict(color="red")) | |
| 476 | assert '"fillColor":"red"' in self._fetch_map_string(m) | |
| 477 | ||
| 478 | m = self.missing.explore("continent", missing_kwds=dict(color="red")) | |
| 479 | assert '"fillColor":"red"' in self._fetch_map_string(m) | |
| 480 | ||
| 481 | def test_categorical_legend(self): | |
| 482 | m = self.world.explore("continent", legend=True) | |
| 483 | out_str = self._fetch_map_string(m) | |
| 484 | assert "#1f77b4'></span>Africa" in out_str | |
| 485 | assert "#ff7f0e'></span>Antarctica" in out_str | |
| 486 | assert "#98df8a'></span>Asia" in out_str | |
| 487 | assert "#9467bd'></span>Europe" in out_str | |
| 488 | assert "#c49c94'></span>NorthAmerica" in out_str | |
| 489 | assert "#7f7f7f'></span>Oceania" in out_str | |
| 490 | assert "#dbdb8d'></span>Sevenseas(openocean)" in out_str | |
| 491 | assert "#9edae5'></span>SouthAmerica" in out_str | |
| 492 | ||
| 493 | m = self.missing.explore( | |
| 494 | "continent", legend=True, missing_kwds={"color": "red"} | |
| 495 | ) | |
| 496 | out_str = self._fetch_map_string(m) | |
| 497 | assert "red'></span>NaN" in out_str | |
| 498 | ||
| 499 | def test_colorbar(self): | |
| 500 | m = self.world.explore("range", legend=True) | |
| 501 | out_str = self._fetch_map_string(m) | |
| 502 | assert "attr(\"id\",'legend')" in out_str | |
| 503 | assert "text('range')" in out_str | |
| 504 | ||
| 505 | m = self.world.explore( | |
| 506 | "range", legend=True, legend_kwds=dict(caption="my_caption") | |
| 507 | ) | |
| 508 | out_str = self._fetch_map_string(m) | |
| 509 | assert "attr(\"id\",'legend')" in out_str | |
| 510 | assert "text('my_caption')" in out_str | |
| 511 | ||
| 512 | m = self.missing.explore("pop_est", legend=True, missing_kwds=dict(color="red")) | |
| 513 | out_str = self._fetch_map_string(m) | |
| 514 | assert "red'></span>NaN" in out_str | |
| 515 | ||
| 516 | # do not scale legend | |
| 517 | m = self.world.explore( | |
| 518 | "pop_est", | |
| 519 | legend=True, | |
| 520 | legend_kwds=dict(scale=False), | |
| 521 | scheme="Headtailbreaks", | |
| 522 | ) | |
| 523 | out_str = self._fetch_map_string(m) | |
| 524 | assert out_str.count("#440154ff") == 100 | |
| 525 | assert out_str.count("#3b528bff") == 100 | |
| 526 | assert out_str.count("#21918cff") == 100 | |
| 527 | assert out_str.count("#5ec962ff") == 100 | |
| 528 | assert out_str.count("#fde725ff") == 100 | |
| 529 | ||
| 530 | # scale legend accorrdingly | |
| 531 | m = self.world.explore( | |
| 532 | "pop_est", | |
| 533 | legend=True, | |
| 534 | scheme="Headtailbreaks", | |
| 535 | ) | |
| 536 | out_str = self._fetch_map_string(m) | |
| 537 | assert out_str.count("#440154ff") == 16 | |
| 538 | assert out_str.count("#3b528bff") == 51 | |
| 539 | assert out_str.count("#21918cff") == 133 | |
| 540 | assert out_str.count("#5ec962ff") == 282 | |
| 541 | assert out_str.count("#fde725ff") == 18 | |
| 542 | ||
| 543 | # discrete cmap | |
| 544 | m = self.world.explore("pop_est", legend=True, cmap="Pastel2") | |
| 545 | out_str = self._fetch_map_string(m) | |
| 546 | ||
| 547 | assert out_str.count("b3e2cdff") == 63 | |
| 548 | assert out_str.count("fdcdacff") == 62 | |
| 549 | assert out_str.count("cbd5e8ff") == 63 | |
| 550 | assert out_str.count("f4cae4ff") == 62 | |
| 551 | assert out_str.count("e6f5c9ff") == 62 | |
| 552 | assert out_str.count("fff2aeff") == 63 | |
| 553 | assert out_str.count("f1e2ccff") == 62 | |
| 554 | assert out_str.count("ccccccff") == 63 | |
| 555 | ||
| 556 | @pytest.mark.skipif(not BRANCA_05, reason="requires branca >= 0.5.0") | |
| 557 | def test_colorbar_max_labels(self): | |
| 558 | # linear | |
| 559 | m = self.world.explore("pop_est", legend_kwds=dict(max_labels=3)) | |
| 560 | out_str = self._fetch_map_string(m) | |
| 561 | ||
| 562 | tick_values = [140.0, 465176713.5921569, 930353287.1843138] | |
| 563 | for tick in tick_values: | |
| 564 | assert str(tick) in out_str | |
| 565 | ||
| 566 | # scheme | |
| 567 | m = self.world.explore( | |
| 568 | "pop_est", scheme="headtailbreaks", legend_kwds=dict(max_labels=3) | |
| 569 | ) | |
| 570 | out_str = self._fetch_map_string(m) | |
| 571 | ||
| 572 | assert "tickValues([140,'',182567501.0,'',1330619341.0,''])" in out_str | |
| 573 | ||
| 574 | # short cmap | |
| 575 | m = self.world.explore("pop_est", legend_kwds=dict(max_labels=3), cmap="tab10") | |
| 576 | out_str = self._fetch_map_string(m) | |
| 577 | ||
| 578 | tick_values = [140.0, 551721192.4, 1103442244.8] | |
| 579 | for tick in tick_values: | |
| 580 | assert str(tick) in out_str | |
| 581 | ||
| 582 | def test_xyzservices_providers(self): | |
| 583 | xyzservices = pytest.importorskip("xyzservices") | |
| 584 | ||
| 585 | m = self.nybb.explore(tiles=xyzservices.providers.CartoDB.PositronNoLabels) | |
| 586 | out_str = self._fetch_map_string(m) | |
| 587 | ||
| 588 | assert ( | |
| 589 | '"https://a.basemaps.cartocdn.com/light_nolabels/{z}/{x}/{y}{r}.png"' | |
| 590 | in out_str | |
| 591 | ) | |
| 592 | assert ( | |
| 593 | 'attribution":"\\u0026copy;\\u003cahref=\\"https://www.openstreetmap.org' | |
| 594 | in out_str | |
| 595 | ) | |
| 596 | assert '"maxNativeZoom":19,"maxZoom":19,"minZoom":0' in out_str | |
| 597 | ||
| 598 | def test_xyzservices_query_name(self): | |
| 599 | pytest.importorskip("xyzservices") | |
| 600 | ||
| 601 | m = self.nybb.explore(tiles="CartoDB Positron No Labels") | |
| 602 | out_str = self._fetch_map_string(m) | |
| 603 | ||
| 604 | assert ( | |
| 605 | '"https://a.basemaps.cartocdn.com/light_nolabels/{z}/{x}/{y}{r}.png"' | |
| 606 | in out_str | |
| 607 | ) | |
| 608 | assert ( | |
| 609 | 'attribution":"\\u0026copy;\\u003cahref=\\"https://www.openstreetmap.org' | |
| 610 | in out_str | |
| 611 | ) | |
| 612 | assert '"maxNativeZoom":19,"maxZoom":19,"minZoom":0' in out_str | |
| 613 | ||
| 614 | def test_linearrings(self): | |
| 615 | rings = self.nybb.explode(index_parts=True).exterior | |
| 616 | m = rings.explore() | |
| 617 | out_str = self._fetch_map_string(m) | |
| 618 | ||
| 619 | assert out_str.count("LineString") == len(rings) | |
| 620 | ||
| 621 | def test_mapclassify_categorical_legend(self): | |
| 622 | m = self.missing.explore( | |
| 623 | column="pop_est", | |
| 624 | legend=True, | |
| 625 | scheme="naturalbreaks", | |
| 626 | missing_kwds=dict(color="red", label="missing"), | |
| 627 | legend_kwds=dict(colorbar=False, interval=True), | |
| 628 | ) | |
| 629 | out_str = self._fetch_map_string(m) | |
| 630 | ||
| 631 | strings = [ | |
| 632 | "[140.00,33986655.00]", | |
| 633 | "(33986655.00,105350020.00]", | |
| 634 | "(105350020.00,207353391.00]", | |
| 635 | "(207353391.00,326625791.00]", | |
| 636 | "(326625791.00,1379302771.00]", | |
| 637 | "missing", | |
| 638 | ] | |
| 639 | for s in strings: | |
| 640 | assert s in out_str | |
| 641 | ||
| 642 | # interval=False | |
| 643 | m = self.missing.explore( | |
| 644 | column="pop_est", | |
| 645 | legend=True, | |
| 646 | scheme="naturalbreaks", | |
| 647 | missing_kwds=dict(color="red", label="missing"), | |
| 648 | legend_kwds=dict(colorbar=False, interval=False), | |
| 649 | ) | |
| 650 | out_str = self._fetch_map_string(m) | |
| 651 | ||
| 652 | strings = [ | |
| 653 | ">140.00,33986655.00", | |
| 654 | ">33986655.00,105350020.00", | |
| 655 | ">105350020.00,207353391.00", | |
| 656 | ">207353391.00,326625791.00", | |
| 657 | ">326625791.00,1379302771.00", | |
| 658 | "missing", | |
| 659 | ] | |
| 660 | for s in strings: | |
| 661 | assert s in out_str | |
| 662 | ||
| 663 | # custom labels | |
| 664 | m = self.world.explore( | |
| 665 | column="pop_est", | |
| 666 | legend=True, | |
| 667 | scheme="naturalbreaks", | |
| 668 | k=5, | |
| 669 | legend_kwds=dict(colorbar=False, labels=["s", "m", "l", "xl", "xxl"]), | |
| 670 | ) | |
| 671 | out_str = self._fetch_map_string(m) | |
| 672 | ||
| 673 | strings = [">s<", ">m<", ">l<", ">xl<", ">xxl<"] | |
| 674 | for s in strings: | |
| 675 | assert s in out_str | |
| 676 | ||
| 677 | # fmt | |
| 678 | m = self.missing.explore( | |
| 679 | column="pop_est", | |
| 680 | legend=True, | |
| 681 | scheme="naturalbreaks", | |
| 682 | missing_kwds=dict(color="red", label="missing"), | |
| 683 | legend_kwds=dict(colorbar=False, fmt="{:.0f}"), | |
| 684 | ) | |
| 685 | out_str = self._fetch_map_string(m) | |
| 686 | ||
| 687 | strings = [ | |
| 688 | ">140,33986655", | |
| 689 | ">33986655,105350020", | |
| 690 | ">105350020,207353391", | |
| 691 | ">207353391,326625791", | |
| 692 | ">326625791,1379302771", | |
| 693 | "missing", | |
| 694 | ] | |
| 695 | for s in strings: | |
| 696 | assert s in out_str | |
| 697 | ||
| 698 | def test_given_m(self): | |
| 699 | "Check that geometry is mapped onto a given folium.Map" | |
| 700 | m = folium.Map() | |
| 701 | self.nybb.explore(m=m, tooltip=False, highlight=False) | |
| 702 | ||
| 703 | out_str = self._fetch_map_string(m) | |
| 704 | ||
| 705 | assert out_str.count("BoroCode") == 5 | |
| 706 | # should not change map settings | |
| 707 | assert m.options["zoom"] == 1 | |
| 708 | ||
| 709 | def test_highlight(self): | |
| 710 | m = self.nybb.explore(highlight=True) | |
| 711 | out_str = self._fetch_map_string(m) | |
| 712 | ||
| 713 | assert '"fillOpacity":0.75' in out_str | |
| 714 | ||
| 715 | m = self.nybb.explore( | |
| 716 | highlight=True, highlight_kwds=dict(fillOpacity=1, color="red") | |
| 717 | ) | |
| 718 | out_str = self._fetch_map_string(m) | |
| 719 | ||
| 720 | assert '{"color":"red","fillOpacity":1}' in out_str | |
| 721 | ||
| 722 | def test_custom_colormaps(self): | |
| 723 | ||
| 724 | step = StepColormap(["green", "yellow", "red"], vmin=0, vmax=100000000) | |
| 725 | ||
| 726 | m = self.world.explore("pop_est", cmap=step, tooltip=["name"], legend=True) | |
| 727 | ||
| 728 | strings = [ | |
| 729 | 'fillColor":"#008000ff"', # Green | |
| 730 | '"fillColor":"#ffff00ff"', # Yellow | |
| 731 | '"fillColor":"#ff0000ff"', # Red | |
| 732 | ] | |
| 733 | ||
| 734 | out_str = self._fetch_map_string(m) | |
| 735 | for s in strings: | |
| 736 | assert s in out_str | |
| 737 | ||
| 738 | assert out_str.count("008000ff") == 306 | |
| 739 | assert out_str.count("ffff00ff") == 187 | |
| 740 | assert out_str.count("ff0000ff") == 190 | |
| 741 | ||
| 742 | # Using custom function colormap | |
| 743 | def my_color_function(field): | |
| 744 | """Maps low values to green and high values to red.""" | |
| 745 | if field > 100000000: | |
| 746 | return "#ff0000" | |
| 747 | else: | |
| 748 | return "#008000" | |
| 749 | ||
| 750 | m = self.world.explore("pop_est", cmap=my_color_function, legend=False) | |
| 751 | ||
| 752 | strings = [ | |
| 753 | '"color":"#ff0000","fillColor":"#ff0000"', | |
| 754 | '"color":"#008000","fillColor":"#008000"', | |
| 755 | ] | |
| 756 | ||
| 757 | for s in strings: | |
| 758 | assert s in self._fetch_map_string(m) | |
| 759 | ||
| 760 | # matplotlib.Colormap | |
| 761 | cmap = colors.ListedColormap(["red", "green", "blue", "white", "black"]) | |
| 762 | ||
| 763 | m = self.nybb.explore("BoroName", cmap=cmap) | |
| 764 | strings = [ | |
| 765 | '"fillColor":"#ff0000"', # Red | |
| 766 | '"fillColor":"#008000"', # Green | |
| 767 | '"fillColor":"#0000ff"', # Blue | |
| 768 | '"fillColor":"#ffffff"', # White | |
| 769 | '"fillColor":"#000000"', # Black | |
| 770 | ] | |
| 771 | ||
| 772 | out_str = self._fetch_map_string(m) | |
| 773 | for s in strings: | |
| 774 | assert s in out_str | |
| 775 | ||
| 776 | def test_multiple_geoseries(self): | |
| 777 | """ | |
| 778 | Additional GeoSeries need to be removed as they cannot be converted to GeoJSON | |
| 779 | """ | |
| 780 | gdf = self.nybb | |
| 781 | gdf["boundary"] = gdf.boundary | |
| 782 | gdf["centroid"] = gdf.centroid | |
| 783 | ||
| 784 | gdf.explore() |
| 22 | 22 | import shapely.geometry |
| 23 | 23 | |
| 24 | 24 | from geopandas.array import GeometryArray, GeometryDtype, from_shapely |
| 25 | from geopandas._compat import ignore_shapely2_warnings | |
| 25 | 26 | |
| 26 | 27 | import pytest |
| 27 | 28 | |
| 47 | 48 | |
| 48 | 49 | def make_data(): |
| 49 | 50 | a = np.empty(100, dtype=object) |
| 50 | a[:] = [shapely.geometry.Point(i, i) for i in range(100)] | |
| 51 | with ignore_shapely2_warnings(): | |
| 52 | a[:] = [shapely.geometry.Point(i, i) for i in range(100)] | |
| 51 | 53 | ga = from_shapely(a) |
| 52 | 54 | return ga |
| 53 | 55 | |
| 298 | 300 | result = np.array(data, dtype=object) |
| 299 | 301 | # expected = np.array(list(data), dtype=object) |
| 300 | 302 | expected = np.empty(len(data), dtype=object) |
| 301 | expected[:] = list(data) | |
| 303 | with ignore_shapely2_warnings(): | |
| 304 | expected[:] = list(data) | |
| 302 | 305 | assert_array_equal(result, expected) |
| 303 | 306 | |
| 304 | 307 | def test_contains(self, data, data_missing): |
| 305 | # overrided due to the inconsistency between | |
| 308 | # overridden due to the inconsistency between | |
| 306 | 309 | # GeometryDtype.na_value = np.nan |
| 307 | 310 | # and None being used as NA in array |
| 308 | 311 | |
| 363 | 366 | |
| 364 | 367 | @pytest.mark.skip("fillna method not supported") |
| 365 | 368 | def test_fillna_series_method(self, data_missing, method): |
| 369 | pass | |
| 370 | ||
| 371 | @pytest.mark.skip("fillna method not supported") | |
| 372 | def test_fillna_no_op_returns_copy(self, data): | |
| 366 | 373 | pass |
| 367 | 374 | |
| 368 | 375 | |
| 394 | 401 | """ |
| 395 | 402 | Fixture for dunder names for common arithmetic operations |
| 396 | 403 | |
| 397 | Adapted to excluse __sub__, as this is implemented as "difference". | |
| 398 | """ | |
| 399 | return request.param | |
| 400 | ||
| 401 | ||
| 404 | Adapted to exclude __sub__, as this is implemented as "difference". | |
| 405 | """ | |
| 406 | return request.param | |
| 407 | ||
| 408 | ||
| 409 | # an inherited test from pandas creates a Series from a list of geometries, which | |
| 410 | # triggers the warning from Shapely, out of control of GeoPandas, so ignoring here | |
| 411 | @pytest.mark.filterwarnings( | |
| 412 | "ignore:The array interface is deprecated and will no longer work in Shapely 2.0" | |
| 413 | ) | |
| 402 | 414 | class TestArithmeticOps(extension_tests.BaseArithmeticOpsTests): |
| 403 | 415 | @pytest.mark.skip(reason="not applicable") |
| 404 | 416 | def test_divmod_series_array(self, data, data_for_twos): |
| 409 | 421 | pass |
| 410 | 422 | |
| 411 | 423 | |
| 424 | # an inherited test from pandas creates a Series from a list of geometries, which | |
| 425 | # triggers the warning from Shapely, out of control of GeoPandas, so ignoring here | |
| 426 | @pytest.mark.filterwarnings( | |
| 427 | "ignore:The array interface is deprecated and will no longer work in Shapely 2.0" | |
| 428 | ) | |
| 412 | 429 | class TestComparisonOps(extension_tests.BaseComparisonOpsTests): |
| 413 | 430 | def _compare_other(self, s, data, op_name, other): |
| 414 | 431 | op = getattr(operator, op_name.strip("_")) |
| 429 | 446 | |
| 430 | 447 | |
| 431 | 448 | class TestMethods(extension_tests.BaseMethodsTests): |
| 432 | @not_yet_implemented | |
| 449 | @no_sorting | |
| 433 | 450 | @pytest.mark.parametrize("dropna", [True, False]) |
| 434 | 451 | def test_value_counts(self, all_data, dropna): |
| 435 | 452 | pass |
| 436 | 453 | |
| 437 | @not_yet_implemented | |
| 454 | @no_sorting | |
| 438 | 455 | def test_value_counts_with_normalize(self, data): |
| 439 | 456 | pass |
| 440 | 457 | |
| 22 | 22 | """ |
| 23 | 23 | |
| 24 | 24 | def __init__(self, *args, **kwargs): |
| 25 | super(ForwardMock, self).__init__(*args, **kwargs) | |
| 25 | super().__init__(*args, **kwargs) | |
| 26 | 26 | self._n = 0.0 |
| 27 | 27 | |
| 28 | 28 | def __call__(self, *args, **kwargs): |
| 29 | 29 | self.return_value = args[0], (self._n, self._n + 0.5) |
| 30 | 30 | self._n += 1 |
| 31 | return super(ForwardMock, self).__call__(*args, **kwargs) | |
| 31 | return super().__call__(*args, **kwargs) | |
| 32 | 32 | |
| 33 | 33 | |
| 34 | 34 | class ReverseMock(mock.MagicMock): |
| 40 | 40 | """ |
| 41 | 41 | |
| 42 | 42 | def __init__(self, *args, **kwargs): |
| 43 | super(ReverseMock, self).__init__(*args, **kwargs) | |
| 43 | super().__init__(*args, **kwargs) | |
| 44 | 44 | self._n = 0 |
| 45 | 45 | |
| 46 | 46 | def __call__(self, *args, **kwargs): |
| 47 | 47 | self.return_value = "address{0}".format(self._n), args[0] |
| 48 | 48 | self._n += 1 |
| 49 | return super(ReverseMock, self).__call__(*args, **kwargs) | |
| 49 | return super().__call__(*args, **kwargs) | |
| 50 | 50 | |
| 51 | 51 | |
| 52 | 52 | @pytest.fixture |
| 133 | 133 | from geopy.exc import GeocoderNotFound |
| 134 | 134 | |
| 135 | 135 | with pytest.raises(GeocoderNotFound): |
| 136 | reverse_geocode(["cambridge, ma"], "badprovider") | |
| 136 | reverse_geocode([Point(0, 0)], "badprovider") | |
| 137 | 137 | |
| 138 | 138 | |
| 139 | 139 | def test_forward(locations, points): |
| 140 | from geopy.geocoders import GeocodeFarm | |
| 140 | from geopy.geocoders import Photon | |
| 141 | 141 | |
| 142 | for provider in ["geocodefarm", GeocodeFarm]: | |
| 143 | with mock.patch("geopy.geocoders.GeocodeFarm.geocode", ForwardMock()) as m: | |
| 142 | for provider in ["photon", Photon]: | |
| 143 | with mock.patch("geopy.geocoders.Photon.geocode", ForwardMock()) as m: | |
| 144 | 144 | g = geocode(locations, provider=provider, timeout=2) |
| 145 | 145 | assert len(locations) == m.call_count |
| 146 | 146 | |
| 154 | 154 | |
| 155 | 155 | |
| 156 | 156 | def test_reverse(locations, points): |
| 157 | from geopy.geocoders import GeocodeFarm | |
| 157 | from geopy.geocoders import Photon | |
| 158 | 158 | |
| 159 | for provider in ["geocodefarm", GeocodeFarm]: | |
| 160 | with mock.patch("geopy.geocoders.GeocodeFarm.reverse", ReverseMock()) as m: | |
| 159 | for provider in ["photon", Photon]: | |
| 160 | with mock.patch("geopy.geocoders.Photon.reverse", ReverseMock()) as m: | |
| 161 | 161 | g = reverse_geocode(points, provider=provider, timeout=2) |
| 162 | 162 | assert len(points) == m.call_count |
| 163 | 163 | |
| 9 | 9 | import pyproj |
| 10 | 10 | from pyproj import CRS |
| 11 | 11 | from pyproj.exceptions import CRSError |
| 12 | from shapely.geometry import Point | |
| 12 | from shapely.geometry import Point, Polygon | |
| 13 | 13 | |
| 14 | 14 | import geopandas |
| 15 | import geopandas._compat as compat | |
| 15 | 16 | from geopandas import GeoDataFrame, GeoSeries, read_file |
| 16 | 17 | from geopandas.array import GeometryArray, GeometryDtype, from_shapely |
| 18 | from geopandas._compat import ignore_shapely2_warnings | |
| 17 | 19 | |
| 18 | 20 | from geopandas.testing import assert_geodataframe_equal, assert_geoseries_equal |
| 19 | 21 | from geopandas.tests.util import PACKAGE_DIR, validate_boro_df |
| 22 | 24 | |
| 23 | 25 | |
| 24 | 26 | PYPROJ_LT_3 = LooseVersion(pyproj.__version__) < LooseVersion("3") |
| 27 | TEST_NEAREST = compat.PYGEOS_GE_010 and compat.USE_PYGEOS | |
| 28 | pandas_133 = pd.__version__ == LooseVersion("1.3.3") | |
| 29 | ||
| 30 | ||
| 31 | @pytest.fixture | |
| 32 | def dfs(request): | |
| 33 | s1 = GeoSeries( | |
| 34 | [ | |
| 35 | Polygon([(0, 0), (2, 0), (2, 2), (0, 2)]), | |
| 36 | Polygon([(2, 2), (4, 2), (4, 4), (2, 4)]), | |
| 37 | ] | |
| 38 | ) | |
| 39 | s2 = GeoSeries( | |
| 40 | [ | |
| 41 | Polygon([(1, 1), (3, 1), (3, 3), (1, 3)]), | |
| 42 | Polygon([(3, 3), (5, 3), (5, 5), (3, 5)]), | |
| 43 | ] | |
| 44 | ) | |
| 45 | df1 = GeoDataFrame({"col1": [1, 2], "geometry": s1}) | |
| 46 | df2 = GeoDataFrame({"col2": [1, 2], "geometry": s2}) | |
| 47 | return df1, df2 | |
| 48 | ||
| 49 | ||
| 50 | @pytest.fixture( | |
| 51 | params=["union", "intersection", "difference", "symmetric_difference", "identity"] | |
| 52 | ) | |
| 53 | def how(request): | |
| 54 | if pandas_133 and request.param in ["symmetric_difference", "identity", "union"]: | |
| 55 | pytest.xfail("Regression in pandas 1.3.3 (GH #2101)") | |
| 56 | return request.param | |
| 25 | 57 | |
| 26 | 58 | |
| 27 | 59 | class TestDataFrame: |
| 366 | 398 | assert len(data["features"]) == 5 |
| 367 | 399 | assert "id" in data["features"][0].keys() |
| 368 | 400 | |
| 401 | @pytest.mark.filterwarnings( | |
| 402 | "ignore:Geometry column does not contain geometry:UserWarning" | |
| 403 | ) | |
| 369 | 404 | def test_to_json_geom_col(self): |
| 370 | 405 | df = self.df.copy() |
| 371 | 406 | df["geom"] = df["geometry"] |
| 456 | 491 | for f in data["features"]: |
| 457 | 492 | assert "id" not in f.keys() |
| 458 | 493 | |
| 494 | def test_to_json_with_duplicate_columns(self): | |
| 495 | df = GeoDataFrame( | |
| 496 | data=[[1, 2, 3]], columns=["a", "b", "a"], geometry=[Point(1, 1)] | |
| 497 | ) | |
| 498 | with pytest.raises( | |
| 499 | ValueError, match="GeoDataFrame cannot contain duplicated column names." | |
| 500 | ): | |
| 501 | df.to_json() | |
| 502 | ||
| 459 | 503 | def test_copy(self): |
| 460 | 504 | df2 = self.df.copy() |
| 461 | 505 | assert type(df2) is GeoDataFrame |
| 484 | 528 | df = GeoDataFrame.from_file(tempfilename) |
| 485 | 529 | assert df.crs == "epsg:2263" |
| 486 | 530 | |
| 531 | def test_to_file_with_duplicate_columns(self): | |
| 532 | df = GeoDataFrame( | |
| 533 | data=[[1, 2, 3]], columns=["a", "b", "a"], geometry=[Point(1, 1)] | |
| 534 | ) | |
| 535 | with pytest.raises( | |
| 536 | ValueError, match="GeoDataFrame cannot contain duplicated column names." | |
| 537 | ): | |
| 538 | tempfilename = os.path.join(self.tempdir, "crs.shp") | |
| 539 | df.to_file(tempfilename) | |
| 540 | ||
| 487 | 541 | def test_bool_index(self): |
| 488 | 542 | # Find boros with 'B' in their name |
| 489 | 543 | df = self.df[self.df["BoroName"].str.contains("B")] |
| 629 | 683 | df = self.df.iloc[:1].copy() |
| 630 | 684 | df.loc[0, "BoroName"] = np.nan |
| 631 | 685 | # when containing missing values |
| 632 | # null: ouput the missing entries as JSON null | |
| 686 | # null: output the missing entries as JSON null | |
| 633 | 687 | result = list(df.iterfeatures(na="null"))[0]["properties"] |
| 634 | 688 | assert result["BoroName"] is None |
| 635 | 689 | # drop: remove the property from the feature. |
| 664 | 718 | # keep |
| 665 | 719 | result = list(df_only_numerical_cols.iterfeatures(na="keep"))[0] |
| 666 | 720 | assert type(result["properties"]["Shape_Leng"]) is float |
| 721 | ||
| 722 | with pytest.raises( | |
| 723 | ValueError, match="GeoDataFrame cannot contain duplicated column names." | |
| 724 | ): | |
| 725 | df_with_duplicate_columns = df[ | |
| 726 | ["Shape_Leng", "Shape_Leng", "Shape_Area", "geometry"] | |
| 727 | ] | |
| 728 | list(df_with_duplicate_columns.iterfeatures()) | |
| 667 | 729 | |
| 668 | 730 | # geometry not set |
| 669 | 731 | df = GeoDataFrame({"values": [0, 1], "geom": [Point(0, 1), Point(1, 0)]}) |
| 743 | 805 | expected_df = pd.DataFrame({"gs0": wkts0, "gs1": wkts1}) |
| 744 | 806 | assert_frame_equal(expected_df, gdf.to_wkt()) |
| 745 | 807 | |
| 808 | @pytest.mark.parametrize("how", ["left", "inner", "right"]) | |
| 809 | @pytest.mark.parametrize("predicate", ["intersects", "within", "contains"]) | |
| 810 | @pytest.mark.skipif( | |
| 811 | not compat.USE_PYGEOS and not compat.HAS_RTREE, | |
| 812 | reason="sjoin needs `rtree` or `pygeos` dependency", | |
| 813 | ) | |
| 814 | def test_sjoin(self, how, predicate): | |
| 815 | """ | |
| 816 | Basic test for availability of the GeoDataFrame method. Other | |
| 817 | sjoin tests are located in /tools/tests/test_sjoin.py | |
| 818 | """ | |
| 819 | left = read_file(geopandas.datasets.get_path("naturalearth_cities")) | |
| 820 | right = read_file(geopandas.datasets.get_path("naturalearth_lowres")) | |
| 821 | ||
| 822 | expected = geopandas.sjoin(left, right, how=how, predicate=predicate) | |
| 823 | result = left.sjoin(right, how=how, predicate=predicate) | |
| 824 | assert_geodataframe_equal(result, expected) | |
| 825 | ||
| 826 | @pytest.mark.parametrize("how", ["left", "inner", "right"]) | |
| 827 | @pytest.mark.parametrize("max_distance", [None, 1]) | |
| 828 | @pytest.mark.parametrize("distance_col", [None, "distance"]) | |
| 829 | @pytest.mark.skipif( | |
| 830 | not TEST_NEAREST, | |
| 831 | reason=( | |
| 832 | "PyGEOS >= 0.10.0" | |
| 833 | " must be installed and activated via the geopandas.compat module to" | |
| 834 | " test sjoin_nearest" | |
| 835 | ), | |
| 836 | ) | |
| 837 | def test_sjoin_nearest(self, how, max_distance, distance_col): | |
| 838 | """ | |
| 839 | Basic test for availability of the GeoDataFrame method. Other | |
| 840 | sjoin tests are located in /tools/tests/test_sjoin.py | |
| 841 | """ | |
| 842 | left = read_file(geopandas.datasets.get_path("naturalearth_cities")) | |
| 843 | right = read_file(geopandas.datasets.get_path("naturalearth_lowres")) | |
| 844 | ||
| 845 | expected = geopandas.sjoin_nearest( | |
| 846 | left, right, how=how, max_distance=max_distance, distance_col=distance_col | |
| 847 | ) | |
| 848 | result = left.sjoin_nearest( | |
| 849 | right, how=how, max_distance=max_distance, distance_col=distance_col | |
| 850 | ) | |
| 851 | assert_geodataframe_equal(result, expected) | |
| 852 | ||
| 853 | @pytest.mark.skip_no_sindex | |
| 854 | def test_clip(self): | |
| 855 | """ | |
| 856 | Basic test for availability of the GeoDataFrame method. Other | |
| 857 | clip tests are located in /tools/tests/test_clip.py | |
| 858 | """ | |
| 859 | left = read_file(geopandas.datasets.get_path("naturalearth_cities")) | |
| 860 | world = read_file(geopandas.datasets.get_path("naturalearth_lowres")) | |
| 861 | south_america = world[world["continent"] == "South America"] | |
| 862 | ||
| 863 | expected = geopandas.clip(left, south_america) | |
| 864 | result = left.clip(south_america) | |
| 865 | assert_geodataframe_equal(result, expected) | |
| 866 | ||
| 867 | @pytest.mark.skip_no_sindex | |
| 868 | def test_overlay(self, dfs, how): | |
| 869 | """ | |
| 870 | Basic test for availability of the GeoDataFrame method. Other | |
| 871 | overlay tests are located in tests/test_overlay.py | |
| 872 | """ | |
| 873 | df1, df2 = dfs | |
| 874 | ||
| 875 | expected = geopandas.overlay(df1, df2, how=how) | |
| 876 | result = df1.overlay(df2, how=how) | |
| 877 | assert_geodataframe_equal(result, expected) | |
| 878 | ||
| 746 | 879 | |
| 747 | 880 | def check_geodataframe(df, geometry_column="geometry"): |
| 748 | 881 | assert isinstance(df, GeoDataFrame) |
| 844 | 977 | "B": np.arange(3.0), |
| 845 | 978 | "geometry": [Point(x, x) for x in range(3)], |
| 846 | 979 | } |
| 847 | a = np.array([data["A"], data["B"], data["geometry"]], dtype=object).T | |
| 980 | with ignore_shapely2_warnings(): | |
| 981 | a = np.array([data["A"], data["B"], data["geometry"]], dtype=object).T | |
| 848 | 982 | |
| 849 | 983 | df = GeoDataFrame(a, columns=["A", "B", "geometry"]) |
| 850 | 984 | check_geodataframe(df) |
| 859 | 993 | "geometry": [Point(x, x) for x in range(3)], |
| 860 | 994 | } |
| 861 | 995 | gpdf = GeoDataFrame(data) |
| 862 | pddf = pd.DataFrame(data) | |
| 996 | with ignore_shapely2_warnings(): | |
| 997 | pddf = pd.DataFrame(data) | |
| 863 | 998 | check_geodataframe(gpdf) |
| 864 | 999 | assert type(pddf) == pd.DataFrame |
| 865 | 1000 | |
| 889 | 1024 | |
| 890 | 1025 | gpdf = GeoDataFrame(data, geometry="other_geom") |
| 891 | 1026 | check_geodataframe(gpdf, "other_geom") |
| 892 | pddf = pd.DataFrame(data) | |
| 1027 | with ignore_shapely2_warnings(): | |
| 1028 | pddf = pd.DataFrame(data) | |
| 893 | 1029 | |
| 894 | 1030 | for df in [gpdf, pddf]: |
| 895 | 1031 | res = GeoDataFrame(df, geometry="other_geom") |
| 896 | 1032 | check_geodataframe(res, "other_geom") |
| 897 | 1033 | |
| 898 | # when passing GeoDataFrame with custom geometry name to constructor | |
| 899 | # an invalid geodataframe is the result TODO is this desired ? | |
| 1034 | # gdf from gdf should preserve active geometry column name | |
| 900 | 1035 | df = GeoDataFrame(gpdf) |
| 901 | with pytest.raises(AttributeError): | |
| 902 | df.geometry | |
| 1036 | check_geodataframe(df, "other_geom") | |
| 903 | 1037 | |
| 904 | 1038 | def test_only_geometry(self): |
| 905 | 1039 | exp = GeoDataFrame( |
| 975 | 1109 | def test_overwrite_geometry(self): |
| 976 | 1110 | # GH602 |
| 977 | 1111 | data = pd.DataFrame({"geometry": [1, 2, 3], "col1": [4, 5, 6]}) |
| 978 | geoms = pd.Series([Point(i, i) for i in range(3)]) | |
| 1112 | with ignore_shapely2_warnings(): | |
| 1113 | geoms = pd.Series([Point(i, i) for i in range(3)]) | |
| 979 | 1114 | # passed geometry kwarg should overwrite geometry column in data |
| 980 | 1115 | res = GeoDataFrame(data, geometry=geoms) |
| 981 | 1116 | assert_geoseries_equal(res.geometry, GeoSeries(geoms)) |
| 982 | 1117 | |
| 1118 | def test_repeat_geo_col(self): | |
| 1119 | df = pd.DataFrame( | |
| 1120 | [ | |
| 1121 | {"geometry": Point(x, y), "geom": Point(x, y)} | |
| 1122 | for x, y in zip(range(3), range(3)) | |
| 1123 | ], | |
| 1124 | ) | |
| 1125 | # explicitly prevent construction of gdf with repeat geometry column names | |
| 1126 | # two columns called "geometry", geom col inferred | |
| 1127 | df2 = df.rename(columns={"geom": "geometry"}) | |
| 1128 | with pytest.raises(ValueError): | |
| 1129 | GeoDataFrame(df2) | |
| 1130 | # ensure case is caught when custom geom column name is used | |
| 1131 | # two columns called "geom", geom col explicit | |
| 1132 | df3 = df.rename(columns={"geometry": "geom"}) | |
| 1133 | with pytest.raises(ValueError): | |
| 1134 | GeoDataFrame(df3, geometry="geom") | |
| 1135 | ||
| 983 | 1136 | |
| 984 | 1137 | def test_geodataframe_crs(): |
| 985 | gdf = GeoDataFrame() | |
| 1138 | gdf = GeoDataFrame(columns=["geometry"]) | |
| 986 | 1139 | gdf.crs = "IGNF:ETRS89UTM28" |
| 987 | 1140 | assert gdf.crs.to_authority() == ("IGNF", "ETRS89UTM28") |
| 1 | 1 | |
| 2 | 2 | import numpy as np |
| 3 | 3 | from numpy.testing import assert_array_equal |
| 4 | from pandas import DataFrame, MultiIndex, Series | |
| 4 | from pandas import DataFrame, Index, MultiIndex, Series | |
| 5 | 5 | |
| 6 | 6 | from shapely.geometry import LinearRing, LineString, MultiPoint, Point, Polygon |
| 7 | 7 | from shapely.geometry.collection import GeometryCollection |
| 103 | 103 | ) |
| 104 | 104 | |
| 105 | 105 | def _test_unary_real(self, op, expected, a): |
| 106 | """ Tests for 'area', 'length', 'is_valid', etc. """ | |
| 106 | """Tests for 'area', 'length', 'is_valid', etc.""" | |
| 107 | 107 | fcmp = assert_series_equal |
| 108 | 108 | self._test_unary(op, expected, a, fcmp) |
| 109 | 109 | |
| 118 | 118 | self._test_unary(op, expected, a, fcmp) |
| 119 | 119 | |
| 120 | 120 | def _test_binary_topological(self, op, expected, a, b, *args, **kwargs): |
| 121 | """ Tests for 'intersection', 'union', 'symmetric_difference', etc. """ | |
| 121 | """Tests for 'intersection', 'union', 'symmetric_difference', etc.""" | |
| 122 | 122 | if isinstance(expected, GeoPandasBase): |
| 123 | 123 | fcmp = assert_geoseries_equal |
| 124 | 124 | else: |
| 228 | 228 | result = getattr(gdf, op) |
| 229 | 229 | fcmp(result, expected) |
| 230 | 230 | |
| 231 | # TODO reenable for all operations once we use pyproj > 2 | |
| 231 | # TODO re-enable for all operations once we use pyproj > 2 | |
| 232 | 232 | # def test_crs_warning(self): |
| 233 | 233 | # # operations on geometries should warn for different CRS |
| 234 | 234 | # no_crs_g3 = self.g3.copy() |
| 244 | 244 | "intersection", self.all_none, self.g1, self.empty |
| 245 | 245 | ) |
| 246 | 246 | |
| 247 | assert len(self.g0.intersection(self.g9, align=True) == 8) | |
| 247 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 248 | assert len(self.g0.intersection(self.g9, align=True) == 8) | |
| 248 | 249 | assert len(self.g0.intersection(self.g9, align=False) == 7) |
| 249 | 250 | |
| 250 | 251 | def test_union_series(self): |
| 251 | 252 | self._test_binary_topological("union", self.sq, self.g1, self.g2) |
| 252 | 253 | |
| 253 | assert len(self.g0.union(self.g9, align=True) == 8) | |
| 254 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 255 | assert len(self.g0.union(self.g9, align=True) == 8) | |
| 254 | 256 | assert len(self.g0.union(self.g9, align=False) == 7) |
| 255 | 257 | |
| 256 | 258 | def test_union_polygon(self): |
| 259 | 261 | def test_symmetric_difference_series(self): |
| 260 | 262 | self._test_binary_topological("symmetric_difference", self.sq, self.g3, self.g4) |
| 261 | 263 | |
| 262 | assert len(self.g0.symmetric_difference(self.g9, align=True) == 8) | |
| 264 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 265 | assert len(self.g0.symmetric_difference(self.g9, align=True) == 8) | |
| 263 | 266 | assert len(self.g0.symmetric_difference(self.g9, align=False) == 7) |
| 264 | 267 | |
| 265 | 268 | def test_symmetric_difference_poly(self): |
| 272 | 275 | expected = GeoSeries([GeometryCollection(), self.t2]) |
| 273 | 276 | self._test_binary_topological("difference", expected, self.g1, self.g2) |
| 274 | 277 | |
| 275 | assert len(self.g0.difference(self.g9, align=True) == 8) | |
| 278 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 279 | assert len(self.g0.difference(self.g9, align=True) == 8) | |
| 276 | 280 | assert len(self.g0.difference(self.g9, align=False) == 7) |
| 277 | 281 | |
| 278 | 282 | def test_difference_poly(self): |
| 289 | 293 | # binary geo empty result with right GeoSeries |
| 290 | 294 | result = GeoSeries([l1]).intersection(GeoSeries([l2])) |
| 291 | 295 | assert_geoseries_equal(result, expected) |
| 292 | # unary geo resulting in emtpy geometry | |
| 296 | # unary geo resulting in empty geometry | |
| 293 | 297 | result = GeoSeries([GeometryCollection()]).convex_hull |
| 294 | 298 | assert_geoseries_equal(result, expected) |
| 295 | 299 | |
| 349 | 353 | |
| 350 | 354 | self._test_unary_topological("unary_union", expected, g) |
| 351 | 355 | |
| 356 | def test_cascaded_union_deprecated(self): | |
| 357 | p1 = self.t1 | |
| 358 | p2 = Polygon([(2, 0), (3, 0), (3, 1)]) | |
| 359 | g = GeoSeries([p1, p2]) | |
| 360 | with pytest.warns( | |
| 361 | FutureWarning, match="The 'cascaded_union' attribute is deprecated" | |
| 362 | ): | |
| 363 | result = g.cascaded_union | |
| 364 | assert result == g.unary_union | |
| 365 | ||
| 352 | 366 | def test_contains(self): |
| 353 | 367 | expected = [True, False, True, False, False, False, False] |
| 354 | 368 | assert_array_dtype_equal(expected, self.g0.contains(self.t1)) |
| 355 | 369 | |
| 356 | 370 | expected = [False, True, True, True, True, True, False, False] |
| 357 | assert_array_dtype_equal(expected, self.g0.contains(self.g9, align=True)) | |
| 371 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 372 | assert_array_dtype_equal(expected, self.g0.contains(self.g9, align=True)) | |
| 358 | 373 | |
| 359 | 374 | expected = [False, False, True, False, False, False, False] |
| 360 | 375 | assert_array_dtype_equal(expected, self.g0.contains(self.g9, align=False)) |
| 378 | 393 | assert_array_dtype_equal(expected, self.crossed_lines.crosses(self.l3)) |
| 379 | 394 | |
| 380 | 395 | expected = [False] * 8 |
| 381 | assert_array_dtype_equal(expected, self.g0.crosses(self.g9, align=True)) | |
| 396 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 397 | assert_array_dtype_equal(expected, self.g0.crosses(self.g9, align=True)) | |
| 382 | 398 | |
| 383 | 399 | expected = [False] * 7 |
| 384 | 400 | assert_array_dtype_equal(expected, self.g0.crosses(self.g9, align=False)) |
| 388 | 404 | assert_array_dtype_equal(expected, self.g0.disjoint(self.t1)) |
| 389 | 405 | |
| 390 | 406 | expected = [False] * 8 |
| 391 | assert_array_dtype_equal(expected, self.g0.disjoint(self.g9, align=True)) | |
| 407 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 408 | assert_array_dtype_equal(expected, self.g0.disjoint(self.g9, align=True)) | |
| 392 | 409 | |
| 393 | 410 | expected = [False, False, False, False, True, False, False] |
| 394 | 411 | assert_array_dtype_equal(expected, self.g0.disjoint(self.g9, align=False)) |
| 425 | 442 | index=range(8), |
| 426 | 443 | ) |
| 427 | 444 | |
| 428 | assert_array_dtype_equal(expected, self.g0.relate(self.g9, align=True)) | |
| 445 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 446 | assert_array_dtype_equal(expected, self.g0.relate(self.g9, align=True)) | |
| 429 | 447 | |
| 430 | 448 | expected = Series( |
| 431 | 449 | [ |
| 451 | 469 | assert_array_dtype_equal(expected, self.g6.distance(self.na_none)) |
| 452 | 470 | |
| 453 | 471 | expected = Series(np.array([np.nan, 0, 0, 0, 0, 0, np.nan, np.nan]), range(8)) |
| 454 | assert_array_dtype_equal(expected, self.g0.distance(self.g9, align=True)) | |
| 472 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 473 | assert_array_dtype_equal(expected, self.g0.distance(self.g9, align=True)) | |
| 455 | 474 | |
| 456 | 475 | val = self.g0.iloc[4].distance(self.g9.iloc[4]) |
| 457 | 476 | expected = Series(np.array([0, 0, 0, 0, val, np.nan, np.nan]), self.g0.index) |
| 478 | 497 | assert_array_dtype_equal(expected, self.g0.intersects(self.empty_poly)) |
| 479 | 498 | |
| 480 | 499 | expected = [False, True, True, True, True, True, False, False] |
| 481 | assert_array_dtype_equal(expected, self.g0.intersects(self.g9, align=True)) | |
| 500 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 501 | assert_array_dtype_equal(expected, self.g0.intersects(self.g9, align=True)) | |
| 482 | 502 | |
| 483 | 503 | expected = [True, True, True, True, False, False, False] |
| 484 | 504 | assert_array_dtype_equal(expected, self.g0.intersects(self.g9, align=False)) |
| 491 | 511 | assert_array_dtype_equal(expected, self.g4.overlaps(self.t1)) |
| 492 | 512 | |
| 493 | 513 | expected = [False] * 8 |
| 494 | assert_array_dtype_equal(expected, self.g0.overlaps(self.g9, align=True)) | |
| 514 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 515 | assert_array_dtype_equal(expected, self.g0.overlaps(self.g9, align=True)) | |
| 495 | 516 | |
| 496 | 517 | expected = [False] * 7 |
| 497 | 518 | assert_array_dtype_equal(expected, self.g0.overlaps(self.g9, align=False)) |
| 501 | 522 | assert_array_dtype_equal(expected, self.g0.touches(self.t1)) |
| 502 | 523 | |
| 503 | 524 | expected = [False] * 8 |
| 504 | assert_array_dtype_equal(expected, self.g0.touches(self.g9, align=True)) | |
| 525 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 526 | assert_array_dtype_equal(expected, self.g0.touches(self.g9, align=True)) | |
| 505 | 527 | |
| 506 | 528 | expected = [True, False, False, True, False, False, False] |
| 507 | 529 | assert_array_dtype_equal(expected, self.g0.touches(self.g9, align=False)) |
| 514 | 536 | assert_array_dtype_equal(expected, self.g0.within(self.sq)) |
| 515 | 537 | |
| 516 | 538 | expected = [False, True, True, True, True, True, False, False] |
| 517 | assert_array_dtype_equal(expected, self.g0.within(self.g9, align=True)) | |
| 539 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 540 | assert_array_dtype_equal(expected, self.g0.within(self.g9, align=True)) | |
| 518 | 541 | |
| 519 | 542 | expected = [False, True, False, False, False, False, False] |
| 520 | 543 | assert_array_dtype_equal(expected, self.g0.within(self.g9, align=False)) |
| 531 | 554 | assert_series_equal(res, exp) |
| 532 | 555 | |
| 533 | 556 | expected = [False, True, True, True, True, True, False, False] |
| 534 | assert_array_dtype_equal(expected, self.g0.covers(self.g9, align=True)) | |
| 557 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 558 | assert_array_dtype_equal(expected, self.g0.covers(self.g9, align=True)) | |
| 535 | 559 | |
| 536 | 560 | expected = [False, False, True, False, False, False, False] |
| 537 | 561 | assert_array_dtype_equal(expected, self.g0.covers(self.g9, align=False)) |
| 551 | 575 | assert_series_equal(res, exp) |
| 552 | 576 | |
| 553 | 577 | expected = [False, True, True, True, True, True, False, False] |
| 554 | assert_array_dtype_equal(expected, self.g0.covered_by(self.g9, align=True)) | |
| 578 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 579 | assert_array_dtype_equal(expected, self.g0.covered_by(self.g9, align=True)) | |
| 555 | 580 | |
| 556 | 581 | expected = [False, True, False, False, False, False, False] |
| 557 | 582 | assert_array_dtype_equal(expected, self.g0.covered_by(self.g9, align=False)) |
| 564 | 589 | expected = Series(np.array([False] * len(self.g1)), self.g1.index) |
| 565 | 590 | self._test_unary_real("is_empty", expected, self.g1) |
| 566 | 591 | |
| 592 | # for is_ring we raise a warning about the value for Polygon changing | |
| 593 | @pytest.mark.filterwarnings("ignore:is_ring:FutureWarning") | |
| 567 | 594 | def test_is_ring(self): |
| 568 | 595 | expected = Series(np.array([True] * len(self.g1)), self.g1.index) |
| 569 | 596 | self._test_unary_real("is_ring", expected, self.g1) |
| 677 | 704 | |
| 678 | 705 | s = GeoSeries([Point(2, 2), Point(0.5, 0.5)], index=[1, 2]) |
| 679 | 706 | expected = Series([np.nan, 2.0, np.nan]) |
| 680 | assert_series_equal(self.g5.project(s), expected) | |
| 707 | with pytest.warns(UserWarning, match="The indices .+ different"): | |
| 708 | assert_series_equal(self.g5.project(s), expected) | |
| 681 | 709 | |
| 682 | 710 | expected = Series([2.0, 0.5], index=self.g5.index) |
| 683 | 711 | assert_series_equal(self.g5.project(s, align=False), expected) |
| 832 | 860 | index=MultiIndex.from_tuples(index, names=expected_index_name), |
| 833 | 861 | crs=4326, |
| 834 | 862 | ) |
| 835 | assert_geoseries_equal(expected, s.explode()) | |
| 863 | with pytest.warns(FutureWarning, match="Currently, index_parts defaults"): | |
| 864 | assert_geoseries_equal(expected, s.explode()) | |
| 836 | 865 | |
| 837 | 866 | @pytest.mark.parametrize("index_name", [None, "test"]) |
| 838 | 867 | def test_explode_geodataframe(self, index_name): |
| 840 | 869 | df = GeoDataFrame({"col": [1, 2], "geometry": s}) |
| 841 | 870 | df.index.name = index_name |
| 842 | 871 | |
| 843 | test_df = df.explode() | |
| 872 | with pytest.warns(FutureWarning, match="Currently, index_parts defaults"): | |
| 873 | test_df = df.explode() | |
| 844 | 874 | |
| 845 | 875 | expected_s = GeoSeries([Point(1, 2), Point(2, 3), Point(5, 5)]) |
| 846 | 876 | expected_df = GeoDataFrame({"col": [1, 1, 2], "geometry": expected_s}) |
| 859 | 889 | df = GeoDataFrame({"level_1": [1, 2], "geometry": s}) |
| 860 | 890 | df.index.name = index_name |
| 861 | 891 | |
| 862 | test_df = df.explode() | |
| 892 | test_df = df.explode(index_parts=True) | |
| 863 | 893 | |
| 864 | 894 | expected_s = GeoSeries([Point(1, 2), Point(2, 3), Point(5, 5)]) |
| 865 | 895 | expected_df = GeoDataFrame({"level_1": [1, 1, 2], "geometry": expected_s}) |
| 871 | 901 | expected_df = expected_df.set_index(expected_index) |
| 872 | 902 | assert_frame_equal(test_df, expected_df) |
| 873 | 903 | |
| 874 | @pytest.mark.skipif( | |
| 875 | not compat.PANDAS_GE_025, | |
| 876 | reason="pandas explode introduced in pandas 0.25", | |
| 877 | ) | |
| 904 | @pytest.mark.parametrize("index_name", [None, "test"]) | |
| 905 | def test_explode_geodataframe_no_multiindex(self, index_name): | |
| 906 | # GH1393 | |
| 907 | s = GeoSeries([MultiPoint([Point(1, 2), Point(2, 3)]), Point(5, 5)]) | |
| 908 | df = GeoDataFrame({"level_1": [1, 2], "geometry": s}) | |
| 909 | df.index.name = index_name | |
| 910 | ||
| 911 | test_df = df.explode(index_parts=False) | |
| 912 | ||
| 913 | expected_s = GeoSeries([Point(1, 2), Point(2, 3), Point(5, 5)]) | |
| 914 | expected_df = GeoDataFrame({"level_1": [1, 1, 2], "geometry": expected_s}) | |
| 915 | ||
| 916 | expected_index = Index([0, 0, 1], name=index_name) | |
| 917 | expected_df = expected_df.set_index(expected_index) | |
| 918 | assert_frame_equal(test_df, expected_df) | |
| 919 | ||
| 878 | 920 | def test_explode_pandas_fallback(self): |
| 879 | 921 | d = { |
| 880 | 922 | "col1": [["name1", "name2"], ["name3", "name4"]], |
| 881 | "geometry": [ | |
| 882 | MultiPoint([(1, 2), (3, 4)]), | |
| 883 | MultiPoint([(2, 1), (0, 0)]), | |
| 884 | ], | |
| 923 | "geometry": [MultiPoint([(1, 2), (3, 4)]), MultiPoint([(2, 1), (0, 0)])], | |
| 885 | 924 | } |
| 886 | 925 | gdf = GeoDataFrame(d, crs=4326) |
| 887 | 926 | expected_df = GeoDataFrame( |
| 913 | 952 | def test_explode_pandas_fallback_ignore_index(self): |
| 914 | 953 | d = { |
| 915 | 954 | "col1": [["name1", "name2"], ["name3", "name4"]], |
| 916 | "geometry": [ | |
| 917 | MultiPoint([(1, 2), (3, 4)]), | |
| 918 | MultiPoint([(2, 1), (0, 0)]), | |
| 919 | ], | |
| 955 | "geometry": [MultiPoint([(1, 2), (3, 4)]), MultiPoint([(2, 1), (0, 0)])], | |
| 920 | 956 | } |
| 921 | 957 | gdf = GeoDataFrame(d, crs=4326) |
| 922 | 958 | expected_df = GeoDataFrame( |
| 940 | 976 | exploded_df = gdf.explode(column="col1", ignore_index=True) |
| 941 | 977 | assert_geodataframe_equal(exploded_df, expected_df) |
| 942 | 978 | |
| 979 | @pytest.mark.parametrize("outer_index", [1, (1, 2), "1"]) | |
| 980 | def test_explode_pandas_multi_index(self, outer_index): | |
| 981 | index = MultiIndex.from_arrays( | |
| 982 | [[outer_index, outer_index, outer_index], [1, 2, 3]], | |
| 983 | names=("first", "second"), | |
| 984 | ) | |
| 985 | df = GeoDataFrame( | |
| 986 | {"vals": [1, 2, 3]}, | |
| 987 | geometry=[MultiPoint([(x, x), (x, 0)]) for x in range(3)], | |
| 988 | index=index, | |
| 989 | ) | |
| 990 | ||
| 991 | test_df = df.explode(index_parts=True) | |
| 992 | ||
| 993 | expected_s = GeoSeries( | |
| 994 | [ | |
| 995 | Point(0, 0), | |
| 996 | Point(0, 0), | |
| 997 | Point(1, 1), | |
| 998 | Point(1, 0), | |
| 999 | Point(2, 2), | |
| 1000 | Point(2, 0), | |
| 1001 | ] | |
| 1002 | ) | |
| 1003 | expected_df = GeoDataFrame({"vals": [1, 1, 2, 2, 3, 3], "geometry": expected_s}) | |
| 1004 | expected_index = MultiIndex.from_tuples( | |
| 1005 | [ | |
| 1006 | (outer_index, *pair) | |
| 1007 | for pair in [(1, 0), (1, 1), (2, 0), (2, 1), (3, 0), (3, 1)] | |
| 1008 | ], | |
| 1009 | names=["first", "second", None], | |
| 1010 | ) | |
| 1011 | expected_df = expected_df.set_index(expected_index) | |
| 1012 | assert_frame_equal(test_df, expected_df) | |
| 1013 | ||
| 1014 | @pytest.mark.parametrize("outer_index", [1, (1, 2), "1"]) | |
| 1015 | def test_explode_pandas_multi_index_false(self, outer_index): | |
| 1016 | index = MultiIndex.from_arrays( | |
| 1017 | [[outer_index, outer_index, outer_index], [1, 2, 3]], | |
| 1018 | names=("first", "second"), | |
| 1019 | ) | |
| 1020 | df = GeoDataFrame( | |
| 1021 | {"vals": [1, 2, 3]}, | |
| 1022 | geometry=[MultiPoint([(x, x), (x, 0)]) for x in range(3)], | |
| 1023 | index=index, | |
| 1024 | ) | |
| 1025 | ||
| 1026 | test_df = df.explode(index_parts=False) | |
| 1027 | ||
| 1028 | expected_s = GeoSeries( | |
| 1029 | [ | |
| 1030 | Point(0, 0), | |
| 1031 | Point(0, 0), | |
| 1032 | Point(1, 1), | |
| 1033 | Point(1, 0), | |
| 1034 | Point(2, 2), | |
| 1035 | Point(2, 0), | |
| 1036 | ] | |
| 1037 | ) | |
| 1038 | expected_df = GeoDataFrame({"vals": [1, 1, 2, 2, 3, 3], "geometry": expected_s}) | |
| 1039 | expected_index = MultiIndex.from_tuples( | |
| 1040 | [ | |
| 1041 | (outer_index, 1), | |
| 1042 | (outer_index, 1), | |
| 1043 | (outer_index, 2), | |
| 1044 | (outer_index, 2), | |
| 1045 | (outer_index, 3), | |
| 1046 | (outer_index, 3), | |
| 1047 | ], | |
| 1048 | names=["first", "second"], | |
| 1049 | ) | |
| 1050 | expected_df = expected_df.set_index(expected_index) | |
| 1051 | assert_frame_equal(test_df, expected_df) | |
| 1052 | ||
| 1053 | @pytest.mark.parametrize("outer_index", [1, (1, 2), "1"]) | |
| 1054 | def test_explode_pandas_multi_index_ignore_index(self, outer_index): | |
| 1055 | index = MultiIndex.from_arrays( | |
| 1056 | [[outer_index, outer_index, outer_index], [1, 2, 3]], | |
| 1057 | names=("first", "second"), | |
| 1058 | ) | |
| 1059 | df = GeoDataFrame( | |
| 1060 | {"vals": [1, 2, 3]}, | |
| 1061 | geometry=[MultiPoint([(x, x), (x, 0)]) for x in range(3)], | |
| 1062 | index=index, | |
| 1063 | ) | |
| 1064 | ||
| 1065 | test_df = df.explode(ignore_index=True) | |
| 1066 | ||
| 1067 | expected_s = GeoSeries( | |
| 1068 | [ | |
| 1069 | Point(0, 0), | |
| 1070 | Point(0, 0), | |
| 1071 | Point(1, 1), | |
| 1072 | Point(1, 0), | |
| 1073 | Point(2, 2), | |
| 1074 | Point(2, 0), | |
| 1075 | ] | |
| 1076 | ) | |
| 1077 | expected_df = GeoDataFrame({"vals": [1, 1, 2, 2, 3, 3], "geometry": expected_s}) | |
| 1078 | expected_index = Index(range(len(expected_df))) | |
| 1079 | expected_df = expected_df.set_index(expected_index) | |
| 1080 | assert_frame_equal(test_df, expected_df) | |
| 1081 | ||
| 1082 | # index_parts is ignored if ignore_index=True | |
| 1083 | test_df = df.explode(ignore_index=True, index_parts=True) | |
| 1084 | assert_frame_equal(test_df, expected_df) | |
| 1085 | ||
| 943 | 1086 | # |
| 944 | 1087 | # Test '&', '|', '^', and '-' |
| 945 | 1088 | # |
| 6 | 6 | import numpy as np |
| 7 | 7 | from numpy.testing import assert_array_equal |
| 8 | 8 | import pandas as pd |
| 9 | from pandas.util.testing import assert_index_equal | |
| 9 | 10 | |
| 10 | 11 | from pyproj import CRS |
| 11 | 12 | from shapely.geometry import ( |
| 18 | 19 | ) |
| 19 | 20 | from shapely.geometry.base import BaseGeometry |
| 20 | 21 | |
| 21 | from geopandas import GeoSeries, GeoDataFrame | |
| 22 | from geopandas._compat import PYPROJ_LT_3 | |
| 22 | from geopandas import GeoSeries, GeoDataFrame, read_file, datasets, clip | |
| 23 | from geopandas._compat import PYPROJ_LT_3, ignore_shapely2_warnings | |
| 23 | 24 | from geopandas.array import GeometryArray, GeometryDtype |
| 24 | 25 | from geopandas.testing import assert_geoseries_equal |
| 25 | 26 | |
| 171 | 172 | assert_series_equal(res, exp) |
| 172 | 173 | |
| 173 | 174 | def test_to_file(self): |
| 174 | """ Test to_file and from_file """ | |
| 175 | """Test to_file and from_file""" | |
| 175 | 176 | tempfilename = os.path.join(self.tempdir, "test.shp") |
| 176 | 177 | self.g3.to_file(tempfilename) |
| 177 | 178 | # Read layer back in? |
| 238 | 239 | # self.na_none.fillna(method='backfill') |
| 239 | 240 | |
| 240 | 241 | def test_coord_slice(self): |
| 241 | """ Test CoordinateSlicer """ | |
| 242 | """Test CoordinateSlicer""" | |
| 242 | 243 | # need some better test cases |
| 243 | 244 | assert geom_equals(self.g3, self.g3.cx[:, :]) |
| 244 | 245 | assert geom_equals(self.g3[[True, False]], self.g3.cx[0.9:, :0.1]) |
| 261 | 262 | |
| 262 | 263 | def test_proj4strings(self): |
| 263 | 264 | # As string |
| 264 | reprojected = self.g3.to_crs("+proj=utm +zone=30N") | |
| 265 | reprojected = self.g3.to_crs("+proj=utm +zone=30") | |
| 265 | 266 | reprojected_back = reprojected.to_crs(epsg=4326) |
| 266 | 267 | assert np.all(self.g3.geom_almost_equals(reprojected_back)) |
| 267 | 268 | |
| 268 | 269 | # As dict |
| 269 | reprojected = self.g3.to_crs({"proj": "utm", "zone": "30N"}) | |
| 270 | reprojected = self.g3.to_crs({"proj": "utm", "zone": "30"}) | |
| 270 | 271 | reprojected_back = reprojected.to_crs(epsg=4326) |
| 271 | 272 | assert np.all(self.g3.geom_almost_equals(reprojected_back)) |
| 272 | 273 | |
| 273 | 274 | # Set to equivalent string, convert, compare to original |
| 274 | 275 | copy = self.g3.copy() |
| 275 | 276 | copy.crs = "epsg:4326" |
| 276 | reprojected = copy.to_crs({"proj": "utm", "zone": "30N"}) | |
| 277 | reprojected = copy.to_crs({"proj": "utm", "zone": "30"}) | |
| 277 | 278 | reprojected_back = reprojected.to_crs(epsg=4326) |
| 278 | 279 | assert np.all(self.g3.geom_almost_equals(reprojected_back)) |
| 279 | 280 | |
| 280 | 281 | # Conversions by different format |
| 281 | reprojected_string = self.g3.to_crs("+proj=utm +zone=30N") | |
| 282 | reprojected_dict = self.g3.to_crs({"proj": "utm", "zone": "30N"}) | |
| 282 | reprojected_string = self.g3.to_crs("+proj=utm +zone=30") | |
| 283 | reprojected_dict = self.g3.to_crs({"proj": "utm", "zone": "30"}) | |
| 283 | 284 | assert np.all(reprojected_string.geom_almost_equals(reprojected_dict)) |
| 284 | 285 | |
| 285 | 286 | def test_from_wkb(self): |
| 321 | 322 | def test_to_wkt(self): |
| 322 | 323 | assert_series_equal(pd.Series([self.t1.wkt, self.sq.wkt]), self.g1.to_wkt()) |
| 323 | 324 | |
| 325 | @pytest.mark.skip_no_sindex | |
| 326 | def test_clip(self): | |
| 327 | left = read_file(datasets.get_path("naturalearth_cities")) | |
| 328 | world = read_file(datasets.get_path("naturalearth_lowres")) | |
| 329 | south_america = world[world["continent"] == "South America"] | |
| 330 | ||
| 331 | expected = clip(left.geometry, south_america) | |
| 332 | result = left.geometry.clip(south_america) | |
| 333 | assert_geoseries_equal(result, expected) | |
| 334 | ||
| 335 | def test_from_xy_points(self): | |
| 336 | x = self.landmarks.x.values | |
| 337 | y = self.landmarks.y.values | |
| 338 | index = self.landmarks.index.tolist() | |
| 339 | crs = self.landmarks.crs | |
| 340 | assert_geoseries_equal( | |
| 341 | self.landmarks, GeoSeries.from_xy(x, y, index=index, crs=crs) | |
| 342 | ) | |
| 343 | assert_geoseries_equal( | |
| 344 | self.landmarks, | |
| 345 | GeoSeries.from_xy(self.landmarks.x, self.landmarks.y, crs=crs), | |
| 346 | ) | |
| 347 | ||
| 348 | def test_from_xy_points_w_z(self): | |
| 349 | index_values = [5, 6, 7] | |
| 350 | x = pd.Series([0, -1, 2], index=index_values) | |
| 351 | y = pd.Series([8, 3, 1], index=index_values) | |
| 352 | z = pd.Series([5, -6, 7], index=index_values) | |
| 353 | expected = GeoSeries( | |
| 354 | [Point(0, 8, 5), Point(-1, 3, -6), Point(2, 1, 7)], index=index_values | |
| 355 | ) | |
| 356 | assert_geoseries_equal(expected, GeoSeries.from_xy(x, y, z)) | |
| 357 | ||
| 358 | def test_from_xy_points_unequal_index(self): | |
| 359 | x = self.landmarks.x | |
| 360 | y = self.landmarks.y | |
| 361 | y.index = -np.arange(len(y)) | |
| 362 | crs = self.landmarks.crs | |
| 363 | assert_geoseries_equal( | |
| 364 | self.landmarks, GeoSeries.from_xy(x, y, index=x.index, crs=crs) | |
| 365 | ) | |
| 366 | unindexed_landmarks = self.landmarks.copy() | |
| 367 | unindexed_landmarks.reset_index(inplace=True, drop=True) | |
| 368 | assert_geoseries_equal( | |
| 369 | unindexed_landmarks, | |
| 370 | GeoSeries.from_xy(x, y, crs=crs), | |
| 371 | ) | |
| 372 | ||
| 373 | def test_from_xy_points_indexless(self): | |
| 374 | x = np.array([0.0, 3.0]) | |
| 375 | y = np.array([2.0, 5.0]) | |
| 376 | z = np.array([-1.0, 4.0]) | |
| 377 | expected = GeoSeries([Point(0, 2, -1), Point(3, 5, 4)]) | |
| 378 | assert_geoseries_equal(expected, GeoSeries.from_xy(x, y, z)) | |
| 379 | ||
| 324 | 380 | |
| 325 | 381 | def test_missing_values_empty_warning(): |
| 326 | 382 | s = GeoSeries([Point(1, 1), None, np.nan, BaseGeometry(), Polygon()]) |
| 354 | 410 | assert len(s.dropna()) == 3 |
| 355 | 411 | |
| 356 | 412 | |
| 413 | def test_isna_empty_geoseries(): | |
| 414 | # ensure that isna() result for emtpy GeoSeries has the correct bool dtype | |
| 415 | s = GeoSeries([]) | |
| 416 | result = s.isna() | |
| 417 | assert_series_equal(result, pd.Series([], dtype="bool")) | |
| 418 | ||
| 419 | ||
| 357 | 420 | def test_geoseries_crs(): |
| 358 | 421 | gs = GeoSeries() |
| 359 | 422 | gs.crs = "IGNF:ETRS89UTM28" |
| 433 | 496 | s = GeoSeries(index=range(3)) |
| 434 | 497 | check_geoseries(s) |
| 435 | 498 | |
| 499 | def test_empty_array(self): | |
| 500 | # with empty data that have an explicit dtype, we use the fallback or | |
| 501 | # not depending on the dtype | |
| 502 | arr = np.array([], dtype="bool") | |
| 503 | ||
| 504 | # dtypes that can never hold geometry-like data | |
| 505 | for arr in [ | |
| 506 | np.array([], dtype="bool"), | |
| 507 | np.array([], dtype="int64"), | |
| 508 | np.array([], dtype="float32"), | |
| 509 | # this gets converted to object dtype by pandas | |
| 510 | # np.array([], dtype="str"), | |
| 511 | ]: | |
| 512 | with pytest.warns(FutureWarning): | |
| 513 | s = GeoSeries(arr) | |
| 514 | assert not isinstance(s, GeoSeries) | |
| 515 | assert type(s) == pd.Series | |
| 516 | ||
| 517 | # dtypes that can potentially hold geometry-like data (object) or | |
| 518 | # can come from empty data (float64) | |
| 519 | for arr in [ | |
| 520 | np.array([], dtype="object"), | |
| 521 | np.array([], dtype="float64"), | |
| 522 | np.array([], dtype="str"), | |
| 523 | ]: | |
| 524 | with pytest.warns(None) as record: | |
| 525 | s = GeoSeries(arr) | |
| 526 | assert not record | |
| 527 | assert isinstance(s, GeoSeries) | |
| 528 | ||
| 436 | 529 | def test_from_series(self): |
| 437 | 530 | shapes = [ |
| 438 | 531 | Polygon([(random.random(), random.random()) for _ in range(3)]) |
| 439 | 532 | for _ in range(10) |
| 440 | 533 | ] |
| 441 | s = pd.Series(shapes, index=list("abcdefghij"), name="foo") | |
| 534 | with ignore_shapely2_warnings(): | |
| 535 | # the warning here is not suppressed by GeoPandas, as this is a pure | |
| 536 | # pandas construction call | |
| 537 | s = pd.Series(shapes, index=list("abcdefghij"), name="foo") | |
| 442 | 538 | g = GeoSeries(s) |
| 443 | 539 | check_geoseries(g) |
| 444 | 540 | |
| 451 | 547 | s = GeoSeries( |
| 452 | 548 | [MultiPoint([(0, 0), (1, 1)]), MultiPoint([(2, 2), (3, 3), (4, 4)])] |
| 453 | 549 | ) |
| 454 | s = s.explode() | |
| 550 | s = s.explode(index_parts=True) | |
| 455 | 551 | df = s.reset_index() |
| 456 | 552 | assert type(df) == GeoDataFrame |
| 553 | ||
| 554 | def test_explode_without_multiindex(self): | |
| 555 | s = GeoSeries( | |
| 556 | [MultiPoint([(0, 0), (1, 1)]), MultiPoint([(2, 2), (3, 3), (4, 4)])] | |
| 557 | ) | |
| 558 | s = s.explode(index_parts=False) | |
| 559 | expected_index = pd.Index([0, 0, 1, 1, 1]) | |
| 560 | assert_index_equal(s.index, expected_index) | |
| 561 | ||
| 562 | def test_explode_ignore_index(self): | |
| 563 | s = GeoSeries( | |
| 564 | [MultiPoint([(0, 0), (1, 1)]), MultiPoint([(2, 2), (3, 3), (4, 4)])] | |
| 565 | ) | |
| 566 | s = s.explode(ignore_index=True) | |
| 567 | expected_index = pd.Index(range(len(s))) | |
| 568 | print(expected_index) | |
| 569 | assert_index_equal(s.index, expected_index) | |
| 570 | ||
| 571 | # index_parts is ignored if ignore_index=True | |
| 572 | s = s.explode(index_parts=True, ignore_index=True) | |
| 573 | assert_index_equal(s.index, expected_index) | |
| 0 | 0 | import pandas as pd |
| 1 | import pytest | |
| 2 | from geopandas.testing import assert_geodataframe_equal | |
| 1 | 3 | |
| 2 | 4 | from shapely.geometry import Point |
| 3 | 5 | |
| 47 | 49 | assert isinstance(res, GeoDataFrame) |
| 48 | 50 | assert isinstance(res.geometry, GeoSeries) |
| 49 | 51 | self._check_metadata(res) |
| 52 | exp = GeoDataFrame(pd.concat([pd.DataFrame(self.gdf), pd.DataFrame(self.gdf)])) | |
| 53 | assert_geodataframe_equal(exp, res) | |
| 54 | # check metadata comes from first gdf | |
| 55 | res4 = pd.concat([self.gdf.set_crs("epsg:4326"), self.gdf], axis=0) | |
| 56 | # Note: this behaviour potentially does not make sense. If geom cols are | |
| 57 | # concatenated but have different CRS, then the CRS will be overridden. | |
| 58 | self._check_metadata(res4, crs="epsg:4326") | |
| 50 | 59 | |
| 51 | 60 | # series |
| 52 | 61 | res = pd.concat([self.gdf.geometry, self.gdf.geometry]) |
| 62 | 71 | assert isinstance(res, GeoDataFrame) |
| 63 | 72 | assert isinstance(res.geometry, GeoSeries) |
| 64 | 73 | self._check_metadata(res) |
| 74 | ||
| 75 | def test_concat_axis1_multiple_geodataframes(self): | |
| 76 | # https://github.com/geopandas/geopandas/issues/1230 | |
| 77 | # Expect that concat should fail gracefully if duplicate column names belonging | |
| 78 | # to geometry columns are introduced. | |
| 79 | expected_err = ( | |
| 80 | "GeoDataFrame does not support multiple columns using the geometry" | |
| 81 | " column name 'geometry'" | |
| 82 | ) | |
| 83 | with pytest.raises(ValueError, match=expected_err): | |
| 84 | pd.concat([self.gdf, self.gdf], axis=1) | |
| 85 | ||
| 86 | # Check case is handled if custom geometry column name is used | |
| 87 | df2 = self.gdf.rename_geometry("geom") | |
| 88 | expected_err2 = ( | |
| 89 | "Concat operation has resulted in multiple columns using the geometry " | |
| 90 | "column name 'geom'." | |
| 91 | ) | |
| 92 | with pytest.raises(ValueError, match=expected_err2): | |
| 93 | pd.concat([df2, df2], axis=1) | |
| 94 | ||
| 95 | # Check that two geometry columns is fine, if they have different names | |
| 96 | res3 = pd.concat([df2.set_crs("epsg:4326"), self.gdf], axis=1) | |
| 97 | # check metadata comes from first df | |
| 98 | self._check_metadata(res3, geometry_column_name="geom", crs="epsg:4326") | |
| 0 | 0 | import os |
| 1 | from distutils.version import LooseVersion | |
| 1 | 2 | |
| 2 | 3 | import pandas as pd |
| 3 | 4 | |
| 6 | 7 | |
| 7 | 8 | import geopandas |
| 8 | 9 | from geopandas import GeoDataFrame, GeoSeries, overlay, read_file |
| 10 | from geopandas import _compat | |
| 9 | 11 | |
| 10 | 12 | from geopandas.testing import assert_geodataframe_equal, assert_geoseries_equal |
| 11 | 13 | import pytest |
| 14 | 16 | |
| 15 | 17 | |
| 16 | 18 | pytestmark = pytest.mark.skip_no_sindex |
| 19 | pandas_133 = pd.__version__ == LooseVersion("1.3.3") | |
| 17 | 20 | |
| 18 | 21 | |
| 19 | 22 | @pytest.fixture |
| 50 | 53 | params=["union", "intersection", "difference", "symmetric_difference", "identity"] |
| 51 | 54 | ) |
| 52 | 55 | def how(request): |
| 56 | if pandas_133 and request.param in ["symmetric_difference", "identity", "union"]: | |
| 57 | pytest.xfail("Regression in pandas 1.3.3 (GH #2101)") | |
| 53 | 58 | return request.param |
| 54 | 59 | |
| 55 | 60 | |
| 182 | 187 | |
| 183 | 188 | # first, check that all bounds and areas are approx equal |
| 184 | 189 | # this is a very rough check for multipolygon equality |
| 190 | if not _compat.PANDAS_GE_11: | |
| 191 | kwargs = dict(check_less_precise=True) | |
| 192 | else: | |
| 193 | kwargs = {} | |
| 185 | 194 | pd.testing.assert_series_equal( |
| 186 | result.geometry.area, expected.geometry.area, check_less_precise=True | |
| 195 | result.geometry.area, expected.geometry.area, **kwargs | |
| 187 | 196 | ) |
| 188 | 197 | pd.testing.assert_frame_equal( |
| 189 | result.geometry.bounds, expected.geometry.bounds, check_less_precise=True | |
| 198 | result.geometry.bounds, expected.geometry.bounds, **kwargs | |
| 190 | 199 | ) |
| 191 | 200 | |
| 192 | 201 | # There are two cases where the multipolygon have a different number |
| 318 | 327 | overlay(df1, df2, how="spandex") |
| 319 | 328 | |
| 320 | 329 | |
| 321 | def test_duplicate_column_name(dfs): | |
| 330 | def test_duplicate_column_name(dfs, how): | |
| 331 | if how == "difference": | |
| 332 | pytest.skip("Difference uses columns from one df only.") | |
| 322 | 333 | df1, df2 = dfs |
| 323 | 334 | df2r = df2.rename(columns={"col2": "col1"}) |
| 324 | res = overlay(df1, df2r, how="union") | |
| 335 | res = overlay(df1, df2r, how=how) | |
| 325 | 336 | assert ("col1_1" in res.columns) and ("col1_2" in res.columns) |
| 326 | 337 | |
| 327 | 338 | |
| 563 | 574 | df1 = read_file(os.path.join(DATA, "geom_type", "df1.geojson")) |
| 564 | 575 | df2 = read_file(os.path.join(DATA, "geom_type", "df2.geojson")) |
| 565 | 576 | |
| 577 | with pytest.warns(UserWarning, match="`keep_geom_type=True` in overlay"): | |
| 578 | intersection = overlay(df1, df2, keep_geom_type=None) | |
| 579 | assert len(intersection) == 1 | |
| 580 | assert (intersection.geom_type == "Polygon").all() | |
| 581 | ||
| 566 | 582 | intersection = overlay(df1, df2, keep_geom_type=True) |
| 567 | 583 | assert len(intersection) == 1 |
| 568 | 584 | assert (intersection.geom_type == "Polygon").all() |
| 570 | 586 | intersection = overlay(df1, df2, keep_geom_type=False) |
| 571 | 587 | assert len(intersection) == 1 |
| 572 | 588 | assert (intersection.geom_type == "GeometryCollection").all() |
| 589 | ||
| 590 | ||
| 591 | def test_keep_geom_type_geometry_collection2(): | |
| 592 | polys1 = [ | |
| 593 | box(0, 0, 1, 1), | |
| 594 | box(1, 1, 3, 3).union(box(1, 3, 5, 5)), | |
| 595 | ] | |
| 596 | ||
| 597 | polys2 = [ | |
| 598 | box(0, 0, 1, 1), | |
| 599 | box(3, 1, 4, 2).union(box(4, 1, 5, 4)), | |
| 600 | ] | |
| 601 | df1 = GeoDataFrame({"left": [0, 1], "geometry": polys1}) | |
| 602 | df2 = GeoDataFrame({"right": [0, 1], "geometry": polys2}) | |
| 603 | ||
| 604 | result1 = overlay(df1, df2, keep_geom_type=True) | |
| 605 | expected1 = GeoDataFrame( | |
| 606 | { | |
| 607 | "left": [0, 1], | |
| 608 | "right": [0, 1], | |
| 609 | "geometry": [box(0, 0, 1, 1), box(4, 3, 5, 4)], | |
| 610 | } | |
| 611 | ) | |
| 612 | assert_geodataframe_equal(result1, expected1) | |
| 613 | ||
| 614 | result1 = overlay(df1, df2, keep_geom_type=False) | |
| 615 | expected1 = GeoDataFrame( | |
| 616 | { | |
| 617 | "left": [0, 1, 1], | |
| 618 | "right": [0, 0, 1], | |
| 619 | "geometry": [ | |
| 620 | box(0, 0, 1, 1), | |
| 621 | Point(1, 1), | |
| 622 | GeometryCollection([box(4, 3, 5, 4), LineString([(3, 1), (3, 2)])]), | |
| 623 | ], | |
| 624 | } | |
| 625 | ) | |
| 626 | assert_geodataframe_equal(result1, expected1) | |
| 573 | 627 | |
| 574 | 628 | |
| 575 | 629 | @pytest.mark.parametrize("make_valid", [True, False]) |
| 591 | 645 | else: |
| 592 | 646 | with pytest.raises(ValueError, match="1 invalid input geometries"): |
| 593 | 647 | overlay(df1, df_bowtie, make_valid=make_valid) |
| 648 | ||
| 649 | ||
| 650 | def test_empty_overlay_return_non_duplicated_columns(): | |
| 651 | ||
| 652 | nybb = geopandas.read_file(geopandas.datasets.get_path("nybb")) | |
| 653 | nybb2 = nybb.copy() | |
| 654 | nybb2.geometry = nybb2.translate(20000000) | |
| 655 | ||
| 656 | result = geopandas.overlay(nybb, nybb2) | |
| 657 | ||
| 658 | assert all(result.columns.isin(nybb.columns)) | |
| 659 | assert len(result.columns) == len(nybb.columns) | |
| 4 | 4 | import pandas as pd |
| 5 | 5 | |
| 6 | 6 | import shapely |
| 7 | from shapely.geometry import Point, GeometryCollection | |
| 7 | from shapely.geometry import Point, GeometryCollection, LineString | |
| 8 | 8 | |
| 9 | 9 | import geopandas |
| 10 | 10 | from geopandas import GeoDataFrame, GeoSeries |
| 45 | 45 | s1 = GeoSeries([p1, p2, None]) |
| 46 | 46 | assert "POINT (10.12346 50.12346)" in repr(s1) |
| 47 | 47 | |
| 48 | # geographic coordinates 4326 | |
| 49 | s3 = GeoSeries([p1, p2], crs=4326) | |
| 50 | assert "POINT (10.12346 50.12346)" in repr(s3) | |
| 51 | ||
| 48 | 52 | # projected coordinates |
| 49 | 53 | p1 = Point(3000.123456789, 3000.123456789) |
| 50 | 54 | p2 = Point(4000.123456789, 4000.123456789) |
| 51 | 55 | s2 = GeoSeries([p1, p2, None]) |
| 52 | 56 | assert "POINT (3000.123 3000.123)" in repr(s2) |
| 57 | ||
| 58 | # projected geographic coordinate | |
| 59 | s4 = GeoSeries([p1, p2], crs=3857) | |
| 60 | assert "POINT (3000.123 3000.123)" in repr(s4) | |
| 53 | 61 | |
| 54 | 62 | geopandas.options.display_precision = 1 |
| 55 | 63 | assert "POINT (10.1 50.1)" in repr(s1) |
| 70 | 78 | def test_repr_empty(): |
| 71 | 79 | # https://github.com/geopandas/geopandas/issues/1195 |
| 72 | 80 | s = GeoSeries([]) |
| 73 | if compat.PANDAS_GE_025: | |
| 74 | # repr with correct name fixed in pandas 0.25 | |
| 75 | assert repr(s) == "GeoSeries([], dtype: geometry)" | |
| 76 | else: | |
| 77 | assert repr(s) == "Series([], dtype: geometry)" | |
| 81 | assert repr(s) == "GeoSeries([], dtype: geometry)" | |
| 78 | 82 | df = GeoDataFrame({"a": [], "geometry": s}) |
| 79 | 83 | assert "Empty GeoDataFrame" in repr(df) |
| 80 | 84 | # https://github.com/geopandas/geopandas/issues/1184 |
| 255 | 259 | res = df.astype(object) |
| 256 | 260 | assert isinstance(res, pd.DataFrame) and not isinstance(res, GeoDataFrame) |
| 257 | 261 | assert res["a"].dtype == object |
| 262 | ||
| 263 | ||
| 264 | @pytest.mark.xfail( | |
| 265 | not compat.PANDAS_GE_10, | |
| 266 | reason="Convert dtypes new in pandas 1.0", | |
| 267 | raises=NotImplementedError, | |
| 268 | ) | |
| 269 | def test_convert_dtypes(df): | |
| 270 | # https://github.com/geopandas/geopandas/issues/1870 | |
| 271 | ||
| 272 | # Test geometry col is first col, first, geom_col_name=geometry | |
| 273 | # (order is important in concat, used internally) | |
| 274 | res1 = df.convert_dtypes() # note res1 done first for pandas < 1 xfail check | |
| 275 | ||
| 276 | expected1 = GeoDataFrame( | |
| 277 | pd.DataFrame(df).convert_dtypes(), crs=df.crs, geometry=df.geometry.name | |
| 278 | ) | |
| 279 | ||
| 280 | # Checking type and metadata are right | |
| 281 | assert_geodataframe_equal(expected1, res1) | |
| 282 | ||
| 283 | # Test geom last, geom_col_name=geometry | |
| 284 | res2 = df[["value1", "value2", "geometry"]].convert_dtypes() | |
| 285 | assert_geodataframe_equal(expected1[["value1", "value2", "geometry"]], res2) | |
| 286 | ||
| 287 | # Test again with crs set and custom geom col name | |
| 288 | df2 = df.set_crs(epsg=4326).rename_geometry("points") | |
| 289 | expected2 = GeoDataFrame( | |
| 290 | pd.DataFrame(df2).convert_dtypes(), crs=df2.crs, geometry=df2.geometry.name | |
| 291 | ) | |
| 292 | res3 = df2.convert_dtypes() | |
| 293 | assert_geodataframe_equal(expected2, res3) | |
| 294 | ||
| 295 | # Test geom last, geom_col=geometry | |
| 296 | res4 = df2[["value1", "value2", "points"]].convert_dtypes() | |
| 297 | assert_geodataframe_equal(expected2[["value1", "value2", "points"]], res4) | |
| 258 | 298 | |
| 259 | 299 | |
| 260 | 300 | def test_to_csv(df): |
| 403 | 443 | assert_array_equal(s.unique(), exp) |
| 404 | 444 | |
| 405 | 445 | |
| 406 | @pytest.mark.xfail | |
| 407 | 446 | def test_value_counts(): |
| 408 | 447 | # each object is considered unique |
| 409 | 448 | s = GeoSeries([Point(0, 0), Point(1, 1), Point(0, 0)]) |
| 410 | 449 | res = s.value_counts() |
| 411 | exp = pd.Series([2, 1], index=[Point(0, 0), Point(1, 1)]) | |
| 450 | with compat.ignore_shapely2_warnings(): | |
| 451 | exp = pd.Series([2, 1], index=[Point(0, 0), Point(1, 1)]) | |
| 412 | 452 | assert_series_equal(res, exp) |
| 453 | # Check crs doesn't make a difference - note it is not kept in output index anyway | |
| 454 | s2 = GeoSeries([Point(0, 0), Point(1, 1), Point(0, 0)], crs="EPSG:4326") | |
| 455 | res2 = s2.value_counts() | |
| 456 | assert_series_equal(res2, exp) | |
| 457 | ||
| 458 | # check mixed geometry | |
| 459 | s3 = GeoSeries([Point(0, 0), LineString([[1, 1], [2, 2]]), Point(0, 0)]) | |
| 460 | res3 = s3.value_counts() | |
| 461 | with compat.ignore_shapely2_warnings(): | |
| 462 | exp3 = pd.Series([2, 1], index=[Point(0, 0), LineString([[1, 1], [2, 2]])]) | |
| 463 | assert_series_equal(res3, exp3) | |
| 464 | ||
| 465 | # check None is handled | |
| 466 | s4 = GeoSeries([Point(0, 0), None, Point(0, 0)]) | |
| 467 | res4 = s4.value_counts(dropna=True) | |
| 468 | with compat.ignore_shapely2_warnings(): | |
| 469 | exp4_dropna = pd.Series([2], index=[Point(0, 0)]) | |
| 470 | assert_series_equal(res4, exp4_dropna) | |
| 471 | with compat.ignore_shapely2_warnings(): | |
| 472 | exp4_keepna = pd.Series([2, 1], index=[Point(0, 0), None]) | |
| 473 | res4_keepna = s4.value_counts(dropna=False) | |
| 474 | assert_series_equal(res4_keepna, exp4_keepna) | |
| 413 | 475 | |
| 414 | 476 | |
| 415 | 477 | @pytest.mark.xfail(strict=False) |
| 453 | 515 | assert_frame_equal(res, exp) |
| 454 | 516 | |
| 455 | 517 | # applying on the geometry column |
| 456 | res = df.groupby("value2")["geometry"].apply(lambda x: x.cascaded_union) | |
| 518 | res = df.groupby("value2")["geometry"].apply(lambda x: x.unary_union) | |
| 457 | 519 | if compat.PANDAS_GE_11: |
| 458 | 520 | exp = GeoSeries( |
| 459 | 521 | [shapely.geometry.MultiPoint([(0, 0), (2, 2)]), Point(1, 1)], |
| 534 | 596 | assert_frame_equal(result, expected) |
| 535 | 597 | |
| 536 | 598 | |
| 599 | def test_apply_preserves_geom_col_name(df): | |
| 600 | df = df.rename_geometry("geom") | |
| 601 | result = df.apply(lambda col: col, axis=0) | |
| 602 | assert result.geometry.name == "geom" | |
| 603 | ||
| 604 | ||
| 537 | 605 | @pytest.mark.skipif(not compat.PANDAS_GE_10, reason="attrs introduced in pandas 1.0") |
| 538 | 606 | def test_preserve_attrs(df): |
| 539 | 607 | # https://github.com/geopandas/geopandas/issues/1654 |
| 549 | 617 | df2 = df.reset_index() |
| 550 | 618 | assert df2.attrs == attrs |
| 551 | 619 | |
| 620 | # https://github.com/geopandas/geopandas/issues/1875 | |
| 621 | df3 = df2.explode(index_parts=True) | |
| 622 | assert df3.attrs == attrs | |
| 623 | ||
| 552 | 624 | |
| 553 | 625 | @pytest.mark.skipif(not compat.PANDAS_GE_12, reason="attrs introduced in pandas 1.0") |
| 554 | 626 | def test_preserve_flags(df): |
| 21 | 21 | from geopandas import GeoDataFrame, GeoSeries, read_file |
| 22 | 22 | from geopandas.datasets import get_path |
| 23 | 23 | import geopandas._compat as compat |
| 24 | from geopandas.plotting import GeoplotAccessor | |
| 24 | 25 | |
| 25 | 26 | import pytest |
| 26 | 27 | |
| 31 | 32 | try: # skipif and importorskip do not work for decorators |
| 32 | 33 | from matplotlib.testing.decorators import check_figures_equal |
| 33 | 34 | |
| 34 | MPL_DECORATORS = True | |
| 35 | if matplotlib.__version__ >= LooseVersion("3.3.0"): | |
| 36 | ||
| 37 | MPL_DECORATORS = True | |
| 38 | else: | |
| 39 | MPL_DECORATORS = False | |
| 35 | 40 | except ImportError: |
| 36 | 41 | MPL_DECORATORS = False |
| 37 | 42 | |
| 510 | 515 | self.df.plot(linestyle=ls, linewidth=1), |
| 511 | 516 | self.df.plot(column="values", linestyle=ls, linewidth=1), |
| 512 | 517 | ]: |
| 513 | np.testing.assert_array_equal(exp_ls, ax.collections[0].get_linestyle()) | |
| 518 | assert exp_ls == ax.collections[0].get_linestyle() | |
| 514 | 519 | |
| 515 | 520 | def test_style_kwargs_linewidth(self): |
| 516 | 521 | # single |
| 544 | 549 | np.linspace(0, 0.0, 1.0, self.N), ax.collections[0].get_alpha() |
| 545 | 550 | ) |
| 546 | 551 | |
| 552 | def test_style_kwargs_path_effects(self): | |
| 553 | from matplotlib.patheffects import withStroke | |
| 554 | ||
| 555 | effects = [withStroke(linewidth=8, foreground="b")] | |
| 556 | ax = self.df.plot(color="orange", path_effects=effects) | |
| 557 | assert ax.collections[0].get_path_effects()[0].__dict__["_gc"] == { | |
| 558 | "linewidth": 8, | |
| 559 | "foreground": "b", | |
| 560 | } | |
| 561 | ||
| 547 | 562 | def test_subplots_norm(self): |
| 548 | 563 | # colors of subplots are the same as for plot (norm is applied) |
| 549 | 564 | cmap = matplotlib.cm.viridis_r |
| 941 | 956 | self.series.plot(linestyles=ls, linewidth=1), |
| 942 | 957 | self.df.plot(linestyles=ls, linewidth=1), |
| 943 | 958 | ]: |
| 944 | np.testing.assert_array_equal(exp_ls, ax.collections[0].get_linestyle()) | |
| 959 | assert exp_ls == ax.collections[0].get_linestyle() | |
| 945 | 960 | |
| 946 | 961 | def test_style_kwargs_linewidth(self): |
| 947 | 962 | # single |
| 1053 | 1068 | pth = get_path("naturalearth_lowres") |
| 1054 | 1069 | cls.df = read_file(pth) |
| 1055 | 1070 | cls.df["NEGATIVES"] = np.linspace(-10, 10, len(cls.df.index)) |
| 1071 | cls.df["low_vals"] = np.linspace(0, 0.3, cls.df.shape[0]) | |
| 1072 | cls.df["mid_vals"] = np.linspace(0.3, 0.7, cls.df.shape[0]) | |
| 1073 | cls.df["high_vals"] = np.linspace(0.7, 1.0, cls.df.shape[0]) | |
| 1074 | cls.df.loc[cls.df.index[:20:2], "high_vals"] = np.nan | |
| 1056 | 1075 | |
| 1057 | 1076 | def test_legend(self): |
| 1058 | 1077 | with warnings.catch_warnings(record=True) as _: # don't print warning |
| 1193 | 1212 | legend_height = _get_ax(fig, "fixed_colorbar").get_position().height |
| 1194 | 1213 | assert abs(plot_height - legend_height) < 1e-6 |
| 1195 | 1214 | |
| 1215 | def test_empty_bins(self): | |
| 1216 | bins = np.arange(1, 11) / 10 | |
| 1217 | ax = self.df.plot( | |
| 1218 | "low_vals", | |
| 1219 | scheme="UserDefined", | |
| 1220 | classification_kwds={"bins": bins}, | |
| 1221 | legend=True, | |
| 1222 | ) | |
| 1223 | expected = np.array( | |
| 1224 | [ | |
| 1225 | [0.281412, 0.155834, 0.469201, 1.0], | |
| 1226 | [0.267004, 0.004874, 0.329415, 1.0], | |
| 1227 | [0.244972, 0.287675, 0.53726, 1.0], | |
| 1228 | ] | |
| 1229 | ) | |
| 1230 | assert all( | |
| 1231 | [ | |
| 1232 | (z == expected).all(axis=1).any() | |
| 1233 | for z in ax.collections[0].get_facecolors() | |
| 1234 | ] | |
| 1235 | ) | |
| 1236 | labels = [ | |
| 1237 | "0.00, 0.10", | |
| 1238 | "0.10, 0.20", | |
| 1239 | "0.20, 0.30", | |
| 1240 | "0.30, 0.40", | |
| 1241 | "0.40, 0.50", | |
| 1242 | "0.50, 0.60", | |
| 1243 | "0.60, 0.70", | |
| 1244 | "0.70, 0.80", | |
| 1245 | "0.80, 0.90", | |
| 1246 | "0.90, 1.00", | |
| 1247 | ] | |
| 1248 | legend = [t.get_text() for t in ax.get_legend().get_texts()] | |
| 1249 | assert labels == legend | |
| 1250 | ||
| 1251 | legend_colors_exp = [ | |
| 1252 | (0.267004, 0.004874, 0.329415, 1.0), | |
| 1253 | (0.281412, 0.155834, 0.469201, 1.0), | |
| 1254 | (0.244972, 0.287675, 0.53726, 1.0), | |
| 1255 | (0.190631, 0.407061, 0.556089, 1.0), | |
| 1256 | (0.147607, 0.511733, 0.557049, 1.0), | |
| 1257 | (0.119699, 0.61849, 0.536347, 1.0), | |
| 1258 | (0.20803, 0.718701, 0.472873, 1.0), | |
| 1259 | (0.430983, 0.808473, 0.346476, 1.0), | |
| 1260 | (0.709898, 0.868751, 0.169257, 1.0), | |
| 1261 | (0.993248, 0.906157, 0.143936, 1.0), | |
| 1262 | ] | |
| 1263 | ||
| 1264 | assert [ | |
| 1265 | line.get_markerfacecolor() for line in ax.get_legend().get_lines() | |
| 1266 | ] == legend_colors_exp | |
| 1267 | ||
| 1268 | ax2 = self.df.plot( | |
| 1269 | "mid_vals", | |
| 1270 | scheme="UserDefined", | |
| 1271 | classification_kwds={"bins": bins}, | |
| 1272 | legend=True, | |
| 1273 | ) | |
| 1274 | expected = np.array( | |
| 1275 | [ | |
| 1276 | [0.244972, 0.287675, 0.53726, 1.0], | |
| 1277 | [0.190631, 0.407061, 0.556089, 1.0], | |
| 1278 | [0.147607, 0.511733, 0.557049, 1.0], | |
| 1279 | [0.119699, 0.61849, 0.536347, 1.0], | |
| 1280 | [0.20803, 0.718701, 0.472873, 1.0], | |
| 1281 | ] | |
| 1282 | ) | |
| 1283 | assert all( | |
| 1284 | [ | |
| 1285 | (z == expected).all(axis=1).any() | |
| 1286 | for z in ax2.collections[0].get_facecolors() | |
| 1287 | ] | |
| 1288 | ) | |
| 1289 | ||
| 1290 | labels = [ | |
| 1291 | "-inf, 0.10", | |
| 1292 | "0.10, 0.20", | |
| 1293 | "0.20, 0.30", | |
| 1294 | "0.30, 0.40", | |
| 1295 | "0.40, 0.50", | |
| 1296 | "0.50, 0.60", | |
| 1297 | "0.60, 0.70", | |
| 1298 | "0.70, 0.80", | |
| 1299 | "0.80, 0.90", | |
| 1300 | "0.90, 1.00", | |
| 1301 | ] | |
| 1302 | legend = [t.get_text() for t in ax2.get_legend().get_texts()] | |
| 1303 | assert labels == legend | |
| 1304 | assert [ | |
| 1305 | line.get_markerfacecolor() for line in ax2.get_legend().get_lines() | |
| 1306 | ] == legend_colors_exp | |
| 1307 | ||
| 1308 | ax3 = self.df.plot( | |
| 1309 | "high_vals", | |
| 1310 | scheme="UserDefined", | |
| 1311 | classification_kwds={"bins": bins}, | |
| 1312 | legend=True, | |
| 1313 | ) | |
| 1314 | expected = np.array( | |
| 1315 | [ | |
| 1316 | [0.709898, 0.868751, 0.169257, 1.0], | |
| 1317 | [0.993248, 0.906157, 0.143936, 1.0], | |
| 1318 | [0.430983, 0.808473, 0.346476, 1.0], | |
| 1319 | ] | |
| 1320 | ) | |
| 1321 | assert all( | |
| 1322 | [ | |
| 1323 | (z == expected).all(axis=1).any() | |
| 1324 | for z in ax3.collections[0].get_facecolors() | |
| 1325 | ] | |
| 1326 | ) | |
| 1327 | ||
| 1328 | legend = [t.get_text() for t in ax3.get_legend().get_texts()] | |
| 1329 | assert labels == legend | |
| 1330 | ||
| 1331 | assert [ | |
| 1332 | line.get_markerfacecolor() for line in ax3.get_legend().get_lines() | |
| 1333 | ] == legend_colors_exp | |
| 1334 | ||
| 1196 | 1335 | |
| 1197 | 1336 | class TestPlotCollections: |
| 1198 | 1337 | def setup_method(self): |
| 1474 | 1613 | ax.cla() |
| 1475 | 1614 | |
| 1476 | 1615 | |
| 1477 | @pytest.mark.skipif(not compat.PANDAS_GE_025, reason="requires pandas > 0.24") | |
| 1478 | 1616 | class TestGeoplotAccessor: |
| 1479 | 1617 | def setup_method(self): |
| 1480 | 1618 | geometries = [Polygon([(0, 0), (1, 0), (1, 1)]), Point(1, 3)] |
| 1498 | 1636 | getattr(self.gdf.plot, kind)(ax=ax_geopandas_2, **kwargs) |
| 1499 | 1637 | |
| 1500 | 1638 | _pandas_kinds = [] |
| 1501 | if compat.PANDAS_GE_025: | |
| 1502 | from geopandas.plotting import GeoplotAccessor | |
| 1503 | ||
| 1504 | _pandas_kinds = GeoplotAccessor._pandas_kinds | |
| 1639 | ||
| 1640 | _pandas_kinds = GeoplotAccessor._pandas_kinds | |
| 1505 | 1641 | |
| 1506 | 1642 | if MPL_DECORATORS: |
| 1507 | 1643 | |
| 1525 | 1661 | kwargs = {"y": "y"} |
| 1526 | 1662 | elif kind in _xy_kinds: |
| 1527 | 1663 | kwargs = {"x": "x", "y": "y"} |
| 1664 | if kind == "hexbin": # increase gridsize to reduce duration | |
| 1665 | kwargs["gridsize"] = 10 | |
| 1528 | 1666 | |
| 1529 | 1667 | self.compare_figures(kind, fig_test, fig_ref, kwargs) |
| 1530 | 1668 | plt.close("all") |
| 1560 | 1698 | polys = GeoSeries([t1, t2], index=list("AB")) |
| 1561 | 1699 | df = GeoDataFrame({"geometry": polys, "values": [0, 1]}) |
| 1562 | 1700 | |
| 1563 | # Test with continous values | |
| 1701 | # Test with continuous values | |
| 1564 | 1702 | ax = df.plot(column="values") |
| 1565 | 1703 | colors = ax.collections[0].get_facecolors() |
| 1566 | 1704 | ax = df.plot(column=df["values"]) |
| 1580 | 1718 | colors_array = ax.collections[0].get_facecolors() |
| 1581 | 1719 | np.testing.assert_array_equal(colors, colors_array) |
| 1582 | 1720 | |
| 1583 | # Check raised error: is df rows number equal to column legth? | |
| 1721 | # Check raised error: is df rows number equal to column length? | |
| 1584 | 1722 | with pytest.raises(ValueError, match="different number of rows"): |
| 1585 | 1723 | ax = df.plot(column=np.array([1, 2, 3])) |
| 1586 | 1724 | |
| 1655 | 1793 | |
| 1656 | 1794 | |
| 1657 | 1795 | def _style_to_vertices(markerstyle): |
| 1658 | """ Converts a markerstyle string to a path. """ | |
| 1796 | """Converts a markerstyle string to a path.""" | |
| 1659 | 1797 | # TODO: Vertices values are twice the actual path; unclear, why. |
| 1660 | 1798 | path = matplotlib.markers.MarkerStyle(markerstyle).get_path() |
| 1661 | 1799 | return path.vertices / 2 |
| 0 | import sys | |
| 0 | from math import sqrt | |
| 1 | 1 | |
| 2 | 2 | from shapely.geometry import ( |
| 3 | 3 | Point, |
| 16 | 16 | import pytest |
| 17 | 17 | import numpy as np |
| 18 | 18 | |
| 19 | ||
| 20 | @pytest.mark.skipif(sys.platform.startswith("win"), reason="fails on AppVeyor") | |
| 19 | if compat.USE_PYGEOS: | |
| 20 | import pygeos | |
| 21 | ||
| 22 | ||
| 21 | 23 | @pytest.mark.skip_no_sindex |
| 22 | 24 | class TestSeriesSindex: |
| 23 | 25 | def test_has_sindex(self): |
| 106 | 108 | assert sliced.sindex is not original_index |
| 107 | 109 | |
| 108 | 110 | |
| 109 | @pytest.mark.skipif(sys.platform.startswith("win"), reason="fails on AppVeyor") | |
| 110 | 111 | @pytest.mark.skip_no_sindex |
| 111 | 112 | class TestFrameSindex: |
| 112 | 113 | def setup_method(self): |
| 161 | 162 | assert geometry_col.sindex is original_index |
| 162 | 163 | |
| 163 | 164 | @pytest.mark.skipif( |
| 164 | not compat.PANDAS_GE_10, reason="Column selection returns a copy on pd<=1.0.0" | |
| 165 | not compat.PANDAS_GE_11, reason="Column selection returns a copy on pd<=1.1.0" | |
| 165 | 166 | ) |
| 166 | 167 | def test_rebuild_on_multiple_col_selection(self): |
| 167 | 168 | """Selecting a subset of columns preserves the index.""" |
| 669 | 670 | ) |
| 670 | 671 | raise e |
| 671 | 672 | |
| 673 | # ------------------------- `nearest` tests ------------------------- # | |
| 674 | @pytest.mark.skipif( | |
| 675 | compat.USE_PYGEOS, | |
| 676 | reason=("RTree supports sindex.nearest with different behaviour"), | |
| 677 | ) | |
| 678 | def test_rtree_nearest_warns(self): | |
| 679 | df = geopandas.GeoDataFrame({"geometry": []}) | |
| 680 | with pytest.warns( | |
| 681 | FutureWarning, match="sindex.nearest using the rtree backend" | |
| 682 | ): | |
| 683 | df.sindex.nearest((0, 0, 1, 1), num_results=2) | |
| 684 | ||
| 685 | @pytest.mark.skipif( | |
| 686 | not (compat.USE_PYGEOS and not compat.PYGEOS_GE_010), | |
| 687 | reason=("PyGEOS < 0.10 does not support sindex.nearest"), | |
| 688 | ) | |
| 689 | def test_pygeos_error(self): | |
| 690 | df = geopandas.GeoDataFrame({"geometry": []}) | |
| 691 | with pytest.raises(NotImplementedError, match="requires pygeos >= 0.10"): | |
| 692 | df.sindex.nearest(None) | |
| 693 | ||
| 694 | @pytest.mark.skipif( | |
| 695 | not (compat.USE_PYGEOS and compat.PYGEOS_GE_010), | |
| 696 | reason=("PyGEOS >= 0.10 is required to test sindex.nearest"), | |
| 697 | ) | |
| 698 | @pytest.mark.parametrize("return_all", [True, False]) | |
| 699 | @pytest.mark.parametrize( | |
| 700 | "geometry,expected", | |
| 701 | [ | |
| 702 | ([0.25, 0.25], [[0], [0]]), | |
| 703 | ([0.75, 0.75], [[0], [1]]), | |
| 704 | ], | |
| 705 | ) | |
| 706 | def test_nearest_single(self, geometry, expected, return_all): | |
| 707 | geoms = pygeos.points(np.arange(10), np.arange(10)) | |
| 708 | df = geopandas.GeoDataFrame({"geometry": geoms}) | |
| 709 | ||
| 710 | p = Point(geometry) | |
| 711 | res = df.sindex.nearest(p, return_all=return_all) | |
| 712 | assert_array_equal(res, expected) | |
| 713 | ||
| 714 | p = pygeos.points(geometry) | |
| 715 | res = df.sindex.nearest(p, return_all=return_all) | |
| 716 | assert_array_equal(res, expected) | |
| 717 | ||
| 718 | @pytest.mark.skipif( | |
| 719 | not compat.USE_PYGEOS or not compat.PYGEOS_GE_010, | |
| 720 | reason=("PyGEOS >= 0.10 is required to test sindex.nearest"), | |
| 721 | ) | |
| 722 | @pytest.mark.parametrize("return_all", [True, False]) | |
| 723 | @pytest.mark.parametrize( | |
| 724 | "geometry,expected", | |
| 725 | [ | |
| 726 | ([(1, 1), (0, 0)], [[0, 1], [1, 0]]), | |
| 727 | ([(1, 1), (0.25, 1)], [[0, 1], [1, 1]]), | |
| 728 | ], | |
| 729 | ) | |
| 730 | def test_nearest_multi(self, geometry, expected, return_all): | |
| 731 | geoms = pygeos.points(np.arange(10), np.arange(10)) | |
| 732 | df = geopandas.GeoDataFrame({"geometry": geoms}) | |
| 733 | ||
| 734 | ps = [Point(p) for p in geometry] | |
| 735 | res = df.sindex.nearest(ps, return_all=return_all) | |
| 736 | assert_array_equal(res, expected) | |
| 737 | ||
| 738 | ps = pygeos.points(geometry) | |
| 739 | res = df.sindex.nearest(ps, return_all=return_all) | |
| 740 | assert_array_equal(res, expected) | |
| 741 | ||
| 742 | s = geopandas.GeoSeries(ps) | |
| 743 | res = df.sindex.nearest(s, return_all=return_all) | |
| 744 | assert_array_equal(res, expected) | |
| 745 | ||
| 746 | x, y = zip(*geometry) | |
| 747 | ga = geopandas.points_from_xy(x, y) | |
| 748 | res = df.sindex.nearest(ga, return_all=return_all) | |
| 749 | assert_array_equal(res, expected) | |
| 750 | ||
| 751 | @pytest.mark.skipif( | |
| 752 | not compat.USE_PYGEOS or not compat.PYGEOS_GE_010, | |
| 753 | reason=("PyGEOS >= 0.10 is required to test sindex.nearest"), | |
| 754 | ) | |
| 755 | @pytest.mark.parametrize("return_all", [True, False]) | |
| 756 | @pytest.mark.parametrize( | |
| 757 | "geometry,expected", | |
| 758 | [ | |
| 759 | (None, [[], []]), | |
| 760 | ([None], [[], []]), | |
| 761 | ], | |
| 762 | ) | |
| 763 | def test_nearest_none(self, geometry, expected, return_all): | |
| 764 | geoms = pygeos.points(np.arange(10), np.arange(10)) | |
| 765 | df = geopandas.GeoDataFrame({"geometry": geoms}) | |
| 766 | ||
| 767 | res = df.sindex.nearest(geometry, return_all=return_all) | |
| 768 | assert_array_equal(res, expected) | |
| 769 | ||
| 770 | @pytest.mark.skipif( | |
| 771 | not compat.USE_PYGEOS or not compat.PYGEOS_GE_010, | |
| 772 | reason=("PyGEOS >= 0.10 is required to test sindex.nearest"), | |
| 773 | ) | |
| 774 | @pytest.mark.parametrize("return_distance", [True, False]) | |
| 775 | @pytest.mark.parametrize( | |
| 776 | "return_all,max_distance,expected", | |
| 777 | [ | |
| 778 | (True, None, ([[0, 0, 1], [0, 1, 5]], [sqrt(0.5), sqrt(0.5), sqrt(50)])), | |
| 779 | (False, None, ([[0, 1], [0, 5]], [sqrt(0.5), sqrt(50)])), | |
| 780 | (True, 1, ([[0, 0], [0, 1]], [sqrt(0.5), sqrt(0.5)])), | |
| 781 | (False, 1, ([[0], [0]], [sqrt(0.5)])), | |
| 782 | ], | |
| 783 | ) | |
| 784 | def test_nearest_max_distance( | |
| 785 | self, expected, max_distance, return_all, return_distance | |
| 786 | ): | |
| 787 | geoms = pygeos.points(np.arange(10), np.arange(10)) | |
| 788 | df = geopandas.GeoDataFrame({"geometry": geoms}) | |
| 789 | ||
| 790 | ps = [Point(0.5, 0.5), Point(0, 10)] | |
| 791 | res = df.sindex.nearest( | |
| 792 | ps, | |
| 793 | return_all=return_all, | |
| 794 | max_distance=max_distance, | |
| 795 | return_distance=return_distance, | |
| 796 | ) | |
| 797 | if return_distance: | |
| 798 | assert_array_equal(res[0], expected[0]) | |
| 799 | assert_array_equal(res[1], expected[1]) | |
| 800 | else: | |
| 801 | assert_array_equal(res, expected[0]) | |
| 802 | ||
| 672 | 803 | # --------------------------- misc tests ---------------------------- # |
| 673 | 804 | |
| 674 | 805 | def test_empty_tree_geometries(self): |
| 128 | 128 | assert_geodataframe_equal(df1, df2, check_crs=False) |
| 129 | 129 | |
| 130 | 130 | assert len(record) == 0 |
| 131 | ||
| 132 | ||
| 133 | def test_almost_equal_but_not_equal(): | |
| 134 | s_origin = GeoSeries([Point(0, 0)]) | |
| 135 | s_almost_origin = GeoSeries([Point(0.0000001, 0)]) | |
| 136 | assert_geoseries_equal(s_origin, s_almost_origin, check_less_precise=True) | |
| 137 | with pytest.raises(AssertionError): | |
| 138 | assert_geoseries_equal(s_origin, s_almost_origin) |
| 0 | 0 | from .crs import explicit_crs_from_epsg |
| 1 | 1 | from .geocoding import geocode, reverse_geocode |
| 2 | 2 | from .overlay import overlay |
| 3 | from .sjoin import sjoin | |
| 3 | from .sjoin import sjoin, sjoin_nearest | |
| 4 | 4 | from .util import collect |
| 5 | 5 | from .clip import clip |
| 6 | 6 | |
| 11 | 11 | "overlay", |
| 12 | 12 | "reverse_geocode", |
| 13 | 13 | "sjoin", |
| 14 | "sjoin_nearest", | |
| 14 | 15 | "clip", |
| 15 | 16 | ] |
| 6 | 6 | """ |
| 7 | 7 | import warnings |
| 8 | 8 | |
| 9 | import numpy as np | |
| 10 | import pandas as pd | |
| 11 | ||
| 12 | 9 | from shapely.geometry import Polygon, MultiPolygon |
| 13 | 10 | |
| 14 | 11 | from geopandas import GeoDataFrame, GeoSeries |
| 15 | 12 | from geopandas.array import _check_crs, _crs_mismatch_warn |
| 16 | 13 | |
| 17 | 14 | |
| 18 | def _clip_points(gdf, poly): | |
| 19 | """Clip point geometry to the polygon extent. | |
| 15 | def _clip_gdf_with_polygon(gdf, poly): | |
| 16 | """Clip geometry to the polygon extent. | |
| 20 | 17 | |
| 21 | Clip an input point GeoDataFrame to the polygon extent of the poly | |
| 22 | parameter. Points that intersect the poly geometry are extracted with | |
| 23 | associated attributes and returned. | |
| 18 | Clip an input GeoDataFrame to the polygon extent of the poly | |
| 19 | parameter. | |
| 24 | 20 | |
| 25 | 21 | Parameters |
| 26 | 22 | ---------- |
| 27 | 23 | gdf : GeoDataFrame, GeoSeries |
| 28 | Composed of point geometry that will be clipped to the poly. | |
| 29 | ||
| 30 | poly : (Multi)Polygon | |
| 31 | Reference geometry used to spatially clip the data. | |
| 32 | ||
| 33 | Returns | |
| 34 | ------- | |
| 35 | GeoDataFrame | |
| 36 | The returned GeoDataFrame is a subset of gdf that intersects | |
| 37 | with poly. | |
| 38 | """ | |
| 39 | return gdf.iloc[gdf.sindex.query(poly, predicate="intersects")] | |
| 40 | ||
| 41 | ||
| 42 | def _clip_line_poly(gdf, poly): | |
| 43 | """Clip line and polygon geometry to the polygon extent. | |
| 44 | ||
| 45 | Clip an input line or polygon to the polygon extent of the poly | |
| 46 | parameter. Parts of Lines or Polygons that intersect the poly geometry are | |
| 47 | extracted with associated attributes and returned. | |
| 48 | ||
| 49 | Parameters | |
| 50 | ---------- | |
| 51 | gdf : GeoDataFrame, GeoSeries | |
| 52 | Line or polygon geometry that is clipped to poly. | |
| 24 | Dataframe to clip. | |
| 53 | 25 | |
| 54 | 26 | poly : (Multi)Polygon |
| 55 | 27 | Reference polygon for clipping. |
| 62 | 34 | """ |
| 63 | 35 | gdf_sub = gdf.iloc[gdf.sindex.query(poly, predicate="intersects")] |
| 64 | 36 | |
| 37 | # For performance reasons points don't need to be intersected with poly | |
| 38 | non_point_mask = gdf_sub.geom_type != "Point" | |
| 39 | ||
| 40 | if not non_point_mask.any(): | |
| 41 | # only points, directly return | |
| 42 | return gdf_sub | |
| 43 | ||
| 65 | 44 | # Clip the data with the polygon |
| 66 | 45 | if isinstance(gdf_sub, GeoDataFrame): |
| 67 | 46 | clipped = gdf_sub.copy() |
| 68 | clipped[gdf.geometry.name] = gdf_sub.intersection(poly) | |
| 47 | clipped.loc[ | |
| 48 | non_point_mask, clipped._geometry_column_name | |
| 49 | ] = gdf_sub.geometry.values[non_point_mask].intersection(poly) | |
| 69 | 50 | else: |
| 70 | 51 | # GeoSeries |
| 71 | clipped = gdf_sub.intersection(poly) | |
| 52 | clipped = gdf_sub.copy() | |
| 53 | clipped[non_point_mask] = gdf_sub.values[non_point_mask].intersection(poly) | |
| 72 | 54 | |
| 73 | 55 | return clipped |
| 74 | 56 | |
| 100 | 82 | GeoDataFrame or GeoSeries |
| 101 | 83 | Vector data (points, lines, polygons) from `gdf` clipped to |
| 102 | 84 | polygon boundary from mask. |
| 85 | ||
| 86 | See also | |
| 87 | -------- | |
| 88 | GeoDataFrame.clip : equivalent GeoDataFrame method | |
| 89 | GeoSeries.clip : equivalent GeoSeries method | |
| 103 | 90 | |
| 104 | 91 | Examples |
| 105 | 92 | -------- |
| 148 | 135 | else: |
| 149 | 136 | poly = mask |
| 150 | 137 | |
| 151 | geom_types = gdf.geometry.type | |
| 152 | poly_idx = np.asarray((geom_types == "Polygon") | (geom_types == "MultiPolygon")) | |
| 153 | line_idx = np.asarray( | |
| 154 | (geom_types == "LineString") | |
| 155 | | (geom_types == "LinearRing") | |
| 156 | | (geom_types == "MultiLineString") | |
| 157 | ) | |
| 158 | point_idx = np.asarray((geom_types == "Point") | (geom_types == "MultiPoint")) | |
| 159 | geomcoll_idx = np.asarray((geom_types == "GeometryCollection")) | |
| 160 | ||
| 161 | if point_idx.any(): | |
| 162 | point_gdf = _clip_points(gdf[point_idx], poly) | |
| 163 | else: | |
| 164 | point_gdf = None | |
| 165 | ||
| 166 | if poly_idx.any(): | |
| 167 | poly_gdf = _clip_line_poly(gdf[poly_idx], poly) | |
| 168 | else: | |
| 169 | poly_gdf = None | |
| 170 | ||
| 171 | if line_idx.any(): | |
| 172 | line_gdf = _clip_line_poly(gdf[line_idx], poly) | |
| 173 | else: | |
| 174 | line_gdf = None | |
| 175 | ||
| 176 | if geomcoll_idx.any(): | |
| 177 | geomcoll_gdf = _clip_line_poly(gdf[geomcoll_idx], poly) | |
| 178 | else: | |
| 179 | geomcoll_gdf = None | |
| 180 | ||
| 181 | order = pd.Series(range(len(gdf)), index=gdf.index) | |
| 182 | concat = pd.concat([point_gdf, line_gdf, poly_gdf, geomcoll_gdf]) | |
| 138 | clipped = _clip_gdf_with_polygon(gdf, poly) | |
| 183 | 139 | |
| 184 | 140 | if keep_geom_type: |
| 185 | geomcoll_concat = (concat.geom_type == "GeometryCollection").any() | |
| 186 | geomcoll_orig = geomcoll_idx.any() | |
| 141 | geomcoll_concat = (clipped.geom_type == "GeometryCollection").any() | |
| 142 | geomcoll_orig = (gdf.geom_type == "GeometryCollection").any() | |
| 187 | 143 | |
| 188 | 144 | new_collection = geomcoll_concat and not geomcoll_orig |
| 189 | 145 | |
| 209 | 165 | # Check how many geometry types are in the clipped GeoDataFrame |
| 210 | 166 | clip_types_total = sum( |
| 211 | 167 | [ |
| 212 | concat.geom_type.isin(polys).any(), | |
| 213 | concat.geom_type.isin(lines).any(), | |
| 214 | concat.geom_type.isin(points).any(), | |
| 168 | clipped.geom_type.isin(polys).any(), | |
| 169 | clipped.geom_type.isin(lines).any(), | |
| 170 | clipped.geom_type.isin(points).any(), | |
| 215 | 171 | ] |
| 216 | 172 | ) |
| 217 | 173 | |
| 225 | 181 | elif new_collection or more_types: |
| 226 | 182 | orig_type = gdf.geom_type.iloc[0] |
| 227 | 183 | if new_collection: |
| 228 | concat = concat.explode() | |
| 184 | clipped = clipped.explode() | |
| 229 | 185 | if orig_type in polys: |
| 230 | concat = concat.loc[concat.geom_type.isin(polys)] | |
| 186 | clipped = clipped.loc[clipped.geom_type.isin(polys)] | |
| 231 | 187 | elif orig_type in lines: |
| 232 | concat = concat.loc[concat.geom_type.isin(lines)] | |
| 188 | clipped = clipped.loc[clipped.geom_type.isin(lines)] | |
| 233 | 189 | |
| 234 | # Return empty GeoDataFrame or GeoSeries if no shapes remain | |
| 235 | if len(concat) == 0: | |
| 236 | return gdf.iloc[:0] | |
| 237 | ||
| 238 | # Preserve the original order of the input | |
| 239 | if isinstance(concat, GeoDataFrame): | |
| 240 | concat["_order"] = order | |
| 241 | return concat.sort_values(by="_order").drop(columns="_order") | |
| 242 | else: | |
| 243 | concat = GeoDataFrame(geometry=concat) | |
| 244 | concat["_order"] = order | |
| 245 | return concat.sort_values(by="_order").geometry | |
| 190 | return clipped | |
| 30 | 30 | strings : list or Series of addresses to geocode |
| 31 | 31 | provider : str or geopy.geocoder |
| 32 | 32 | Specifies geocoding service to use. If none is provided, |
| 33 | will use 'geocodefarm' with a rate limit applied (see the geocodefarm | |
| 34 | terms of service at: | |
| 35 | https://geocode.farm/geocoding/free-api-documentation/ ). | |
| 33 | will use 'photon' (see the Photon's terms of service at: | |
| 34 | https://photon.komoot.io). | |
| 36 | 35 | |
| 37 | 36 | Either the string name used by geopy (as specified in |
| 38 | 37 | geopy.geocoders.SERVICE_TO_GEOCODER) or a geopy Geocoder instance |
| 39 | (e.g., geopy.geocoders.GeocodeFarm) may be used. | |
| 38 | (e.g., geopy.geocoders.Photon) may be used. | |
| 40 | 39 | |
| 41 | 40 | Some providers require additional arguments such as access keys |
| 42 | 41 | See each geocoder's specific parameters in geopy.geocoders |
| 61 | 60 | """ |
| 62 | 61 | |
| 63 | 62 | if provider is None: |
| 64 | # https://geocode.farm/geocoding/free-api-documentation/ | |
| 65 | provider = "geocodefarm" | |
| 66 | throttle_time = 0.25 | |
| 67 | else: | |
| 68 | throttle_time = _get_throttle_time(provider) | |
| 63 | provider = "photon" | |
| 64 | throttle_time = _get_throttle_time(provider) | |
| 69 | 65 | |
| 70 | 66 | return _query(strings, True, provider, throttle_time, **kwargs) |
| 71 | 67 | |
| 84 | 80 | y coordinate is latitude |
| 85 | 81 | provider : str or geopy.geocoder (opt) |
| 86 | 82 | Specifies geocoding service to use. If none is provided, |
| 87 | will use 'geocodefarm' with a rate limit applied (see the geocodefarm | |
| 88 | terms of service at: | |
| 89 | https://geocode.farm/geocoding/free-api-documentation/ ). | |
| 83 | will use 'photon' (see the Photon's terms of service at: | |
| 84 | https://photon.komoot.io). | |
| 90 | 85 | |
| 91 | 86 | Either the string name used by geopy (as specified in |
| 92 | 87 | geopy.geocoders.SERVICE_TO_GEOCODER) or a geopy Geocoder instance |
| 93 | (e.g., geopy.geocoders.GeocodeFarm) may be used. | |
| 88 | (e.g., geopy.geocoders.Photon) may be used. | |
| 94 | 89 | |
| 95 | 90 | Some providers require additional arguments such as access keys |
| 96 | 91 | See each geocoder's specific parameters in geopy.geocoders |
| 116 | 111 | """ |
| 117 | 112 | |
| 118 | 113 | if provider is None: |
| 119 | # https://geocode.farm/geocoding/free-api-documentation/ | |
| 120 | provider = "geocodefarm" | |
| 121 | throttle_time = 0.25 | |
| 122 | else: | |
| 123 | throttle_time = _get_throttle_time(provider) | |
| 114 | provider = "photon" | |
| 115 | throttle_time = _get_throttle_time(provider) | |
| 124 | 116 | |
| 125 | 117 | return _query(points, False, provider, throttle_time, **kwargs) |
| 126 | 118 | |
| 130 | 122 | from geopy.geocoders.base import GeocoderQueryError |
| 131 | 123 | from geopy.geocoders import get_geocoder_for_service |
| 132 | 124 | |
| 133 | if not isinstance(data, pd.Series): | |
| 134 | data = pd.Series(data) | |
| 125 | if forward: | |
| 126 | if not isinstance(data, pd.Series): | |
| 127 | data = pd.Series(data) | |
| 128 | else: | |
| 129 | if not isinstance(data, geopandas.GeoSeries): | |
| 130 | data = geopandas.GeoSeries(data) | |
| 135 | 131 | |
| 136 | 132 | if isinstance(provider, str): |
| 137 | 133 | provider = get_geocoder_for_service(provider) |
| 180 | 180 | |
| 181 | 181 | >>> geopandas.overlay(df1, df2, how='union') |
| 182 | 182 | df1_data df2_data geometry |
| 183 | 0 1.0 1.0 POLYGON ((1.00000 2.00000, 2.00000 2.00000, 2.... | |
| 184 | 1 2.0 1.0 POLYGON ((3.00000 2.00000, 2.00000 2.00000, 2.... | |
| 185 | 2 2.0 2.0 POLYGON ((3.00000 4.00000, 4.00000 4.00000, 4.... | |
| 186 | 3 1.0 NaN POLYGON ((2.00000 1.00000, 2.00000 0.00000, 0.... | |
| 183 | 0 1.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 184 | 1 2.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 185 | 2 2.0 2.0 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 186 | 3 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 187 | 187 | 4 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... |
| 188 | 188 | 5 NaN 1.0 MULTIPOLYGON (((2.00000 2.00000, 3.00000 2.000... |
| 189 | 6 NaN 2.0 POLYGON ((3.00000 4.00000, 3.00000 5.00000, 5.... | |
| 189 | 6 NaN 2.0 POLYGON ((3.00000 5.00000, 5.00000 5.00000, 5.... | |
| 190 | 190 | |
| 191 | 191 | >>> geopandas.overlay(df1, df2, how='intersection') |
| 192 | 192 | df1_data df2_data geometry |
| 193 | 0 1 1 POLYGON ((1.00000 2.00000, 2.00000 2.00000, 2.... | |
| 194 | 1 2 1 POLYGON ((3.00000 2.00000, 2.00000 2.00000, 2.... | |
| 195 | 2 2 2 POLYGON ((3.00000 4.00000, 4.00000 4.00000, 4.... | |
| 193 | 0 1 1 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 194 | 1 2 1 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 195 | 2 2 2 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 196 | 196 | |
| 197 | 197 | >>> geopandas.overlay(df1, df2, how='symmetric_difference') |
| 198 | 198 | df1_data df2_data geometry |
| 199 | 0 1.0 NaN POLYGON ((2.00000 1.00000, 2.00000 0.00000, 0.... | |
| 199 | 0 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 200 | 200 | 1 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... |
| 201 | 201 | 2 NaN 1.0 MULTIPOLYGON (((2.00000 2.00000, 3.00000 2.000... |
| 202 | 3 NaN 2.0 POLYGON ((3.00000 4.00000, 3.00000 5.00000, 5.... | |
| 202 | 3 NaN 2.0 POLYGON ((3.00000 5.00000, 5.00000 5.00000, 5.... | |
| 203 | 203 | |
| 204 | 204 | >>> geopandas.overlay(df1, df2, how='difference') |
| 205 | geometry df1_data | |
| 206 | 0 POLYGON ((2.00000 1.00000, 2.00000 0.00000, 0.... 1 | |
| 207 | 1 MULTIPOLYGON (((2.00000 3.00000, 2.00000 4.000... 2 | |
| 205 | geometry df1_data | |
| 206 | 0 POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... 1 | |
| 207 | 1 MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... 2 | |
| 208 | 208 | |
| 209 | 209 | >>> geopandas.overlay(df1, df2, how='identity') |
| 210 | 210 | df1_data df2_data geometry |
| 211 | 0 1.0 1.0 POLYGON ((1.00000 2.00000, 2.00000 2.00000, 2.... | |
| 212 | 1 2.0 1.0 POLYGON ((3.00000 2.00000, 2.00000 2.00000, 2.... | |
| 213 | 2 2.0 2.0 POLYGON ((3.00000 4.00000, 4.00000 4.00000, 4.... | |
| 214 | 3 1.0 NaN POLYGON ((2.00000 1.00000, 2.00000 0.00000, 0.... | |
| 211 | 0 1.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 1.00000, 1.... | |
| 212 | 1 2.0 1.0 POLYGON ((2.00000 2.00000, 2.00000 3.00000, 3.... | |
| 213 | 2 2.0 2.0 POLYGON ((4.00000 4.00000, 4.00000 3.00000, 3.... | |
| 214 | 3 1.0 NaN POLYGON ((2.00000 0.00000, 0.00000 0.00000, 0.... | |
| 215 | 215 | 4 2.0 NaN MULTIPOLYGON (((3.00000 3.00000, 4.00000 3.000... |
| 216 | 216 | |
| 217 | 217 | See also |
| 218 | 218 | -------- |
| 219 | 219 | sjoin : spatial join |
| 220 | GeoDataFrame.overlay : equivalent method | |
| 220 | 221 | |
| 221 | 222 | Notes |
| 222 | 223 | ------ |
| 262 | 263 | raise NotImplementedError( |
| 263 | 264 | "df{} contains mixed geometry types.".format(i + 1) |
| 264 | 265 | ) |
| 266 | ||
| 267 | box_gdf1 = df1.total_bounds | |
| 268 | box_gdf2 = df2.total_bounds | |
| 269 | ||
| 270 | if not ( | |
| 271 | ((box_gdf1[0] <= box_gdf2[2]) and (box_gdf2[0] <= box_gdf1[2])) | |
| 272 | and ((box_gdf1[1] <= box_gdf2[3]) and (box_gdf2[1] <= box_gdf1[3])) | |
| 273 | ): | |
| 274 | return GeoDataFrame( | |
| 275 | [], | |
| 276 | columns=list( | |
| 277 | set( | |
| 278 | df1.drop(df1.geometry.name, axis=1).columns.to_list() | |
| 279 | + df2.drop(df2.geometry.name, axis=1).columns.to_list() | |
| 280 | ) | |
| 281 | ) | |
| 282 | + ["geometry"], | |
| 283 | ) | |
| 265 | 284 | |
| 266 | 285 | # Computations |
| 267 | 286 | def _make_valid(df): |
| 283 | 302 | df1 = _make_valid(df1) |
| 284 | 303 | df2 = _make_valid(df2) |
| 285 | 304 | |
| 286 | with warnings.catch_warnings(): # CRS checked above, supress array-level warning | |
| 305 | with warnings.catch_warnings(): # CRS checked above, suppress array-level warning | |
| 287 | 306 | warnings.filterwarnings("ignore", message="CRS mismatch between the CRS") |
| 288 | 307 | if how == "difference": |
| 289 | 308 | return _overlay_difference(df1, df2) |
| 298 | 317 | result = dfunion[dfunion["__idx1"].notnull()].copy() |
| 299 | 318 | |
| 300 | 319 | if keep_geom_type: |
| 301 | key_order = result.keys() | |
| 302 | exploded = result.reset_index(drop=True).explode() | |
| 303 | exploded = exploded.reset_index(level=0) | |
| 304 | ||
| 320 | geom_type = df1.geom_type.iloc[0] | |
| 321 | ||
| 322 | # First we filter the geometry types inside GeometryCollections objects | |
| 323 | # (e.g. GeometryCollection([polygon, point]) -> polygon) | |
| 324 | # we do this separately on only the relevant rows, as this is an expensive | |
| 325 | # operation (an expensive no-op for geometry types other than collections) | |
| 326 | is_collection = result.geom_type == "GeometryCollection" | |
| 327 | if is_collection.any(): | |
| 328 | geom_col = result._geometry_column_name | |
| 329 | collections = result[[geom_col]][is_collection] | |
| 330 | ||
| 331 | exploded = collections.reset_index(drop=True).explode(index_parts=True) | |
| 332 | exploded = exploded.reset_index(level=0) | |
| 333 | ||
| 334 | orig_num_geoms_exploded = exploded.shape[0] | |
| 335 | if geom_type in polys: | |
| 336 | exploded = exploded.loc[exploded.geom_type.isin(polys)] | |
| 337 | elif geom_type in lines: | |
| 338 | exploded = exploded.loc[exploded.geom_type.isin(lines)] | |
| 339 | elif geom_type in points: | |
| 340 | exploded = exploded.loc[exploded.geom_type.isin(points)] | |
| 341 | else: | |
| 342 | raise TypeError( | |
| 343 | "`keep_geom_type` does not support {}.".format(geom_type) | |
| 344 | ) | |
| 345 | num_dropped_collection = orig_num_geoms_exploded - exploded.shape[0] | |
| 346 | ||
| 347 | # level_0 created with above reset_index operation | |
| 348 | # and represents the original geometry collections | |
| 349 | # TODO avoiding dissolve to call unary_union in this case could further | |
| 350 | # improve performance (we only need to collect geometries in their | |
| 351 | # respective Multi version) | |
| 352 | dissolved = exploded.dissolve(by="level_0") | |
| 353 | result.loc[is_collection, geom_col] = dissolved[geom_col].values | |
| 354 | else: | |
| 355 | num_dropped_collection = 0 | |
| 356 | ||
| 357 | # Now we filter all geometries (in theory we don't need to do this | |
| 358 | # again for the rows handled above for GeometryCollections, but filtering | |
| 359 | # them out is probably more expensive as simply including them when this | |
| 360 | # is typically about only a few rows) | |
| 305 | 361 | orig_num_geoms = result.shape[0] |
| 306 | geom_type = df1.geom_type.iloc[0] | |
| 307 | 362 | if geom_type in polys: |
| 308 | exploded = exploded.loc[exploded.geom_type.isin(polys)] | |
| 363 | result = result.loc[result.geom_type.isin(polys)] | |
| 309 | 364 | elif geom_type in lines: |
| 310 | exploded = exploded.loc[exploded.geom_type.isin(lines)] | |
| 365 | result = result.loc[result.geom_type.isin(lines)] | |
| 311 | 366 | elif geom_type in points: |
| 312 | exploded = exploded.loc[exploded.geom_type.isin(points)] | |
| 367 | result = result.loc[result.geom_type.isin(points)] | |
| 313 | 368 | else: |
| 314 | 369 | raise TypeError("`keep_geom_type` does not support {}.".format(geom_type)) |
| 315 | ||
| 316 | # level_0 created with above reset_index operation | |
| 317 | # and represents the original geometry collections | |
| 318 | result = exploded.dissolve(by="level_0")[key_order] | |
| 319 | ||
| 320 | if (result.shape[0] != orig_num_geoms) and keep_geom_type_warning: | |
| 321 | num_dropped = orig_num_geoms - result.shape[0] | |
| 370 | num_dropped = orig_num_geoms - result.shape[0] | |
| 371 | ||
| 372 | if (num_dropped > 0 or num_dropped_collection > 0) and keep_geom_type_warning: | |
| 322 | 373 | warnings.warn( |
| 323 | 374 | "`keep_geom_type=True` in overlay resulted in {} dropped " |
| 324 | 375 | "geometries of different geometry types than df1 has. " |
| 325 | 376 | "Set `keep_geom_type=False` to retain all " |
| 326 | "geometries".format(num_dropped), | |
| 377 | "geometries".format(num_dropped + num_dropped_collection), | |
| 327 | 378 | UserWarning, |
| 328 | 379 | stacklevel=2, |
| 329 | 380 | ) |
| 0 | from typing import Optional | |
| 0 | 1 | import warnings |
| 1 | 2 | |
| 3 | import numpy as np | |
| 2 | 4 | import pandas as pd |
| 3 | 5 | |
| 4 | 6 | from geopandas import GeoDataFrame |
| 7 | from geopandas import _compat as compat | |
| 5 | 8 | from geopandas.array import _check_crs, _crs_mismatch_warn |
| 6 | 9 | |
| 7 | 10 | |
| 8 | 11 | def sjoin( |
| 9 | left_df, right_df, how="inner", op="intersects", lsuffix="left", rsuffix="right" | |
| 12 | left_df, | |
| 13 | right_df, | |
| 14 | how="inner", | |
| 15 | predicate="intersects", | |
| 16 | lsuffix="left", | |
| 17 | rsuffix="right", | |
| 18 | **kwargs, | |
| 10 | 19 | ): |
| 11 | 20 | """Spatial join of two GeoDataFrames. |
| 12 | 21 | |
| 23 | 32 | * 'right': use keys from right_df; retain only right_df geometry column |
| 24 | 33 | * 'inner': use intersection of keys from both dfs; retain only |
| 25 | 34 | left_df geometry column |
| 26 | op : string, default 'intersects' | |
| 35 | predicate : string, default 'intersects' | |
| 27 | 36 | Binary predicate. Valid values are determined by the spatial index used. |
| 28 | 37 | You can check the valid values in left_df or right_df as |
| 29 | 38 | ``left_df.sindex.valid_query_predicates`` or |
| 30 | 39 | ``right_df.sindex.valid_query_predicates`` |
| 40 | Replaces deprecated ``op`` parameter. | |
| 31 | 41 | lsuffix : string, default 'left' |
| 32 | 42 | Suffix to apply to overlapping column names (left GeoDataFrame). |
| 33 | 43 | rsuffix : string, default 'right' |
| 77 | 87 | See also |
| 78 | 88 | -------- |
| 79 | 89 | overlay : overlay operation resulting in a new geometry |
| 90 | GeoDataFrame.sjoin : equivalent method | |
| 80 | 91 | |
| 81 | 92 | Notes |
| 82 | 93 | ------ |
| 83 | 94 | Every operation in GeoPandas is planar, i.e. the potential third |
| 84 | 95 | dimension is not taken into account. |
| 85 | 96 | """ |
| 97 | if "op" in kwargs: | |
| 98 | op = kwargs.pop("op") | |
| 99 | deprecation_message = ( | |
| 100 | "The `op` parameter is deprecated and will be removed" | |
| 101 | " in a future release. Please use the `predicate` parameter" | |
| 102 | " instead." | |
| 103 | ) | |
| 104 | if predicate != "intersects" and op != predicate: | |
| 105 | override_message = ( | |
| 106 | "A non-default value for `predicate` was passed" | |
| 107 | f' (got `predicate="{predicate}"`' | |
| 108 | f' in combination with `op="{op}"`).' | |
| 109 | " The value of `predicate` will be overriden by the value of `op`," | |
| 110 | " , which may result in unexpected behavior." | |
| 111 | f"\n{deprecation_message}" | |
| 112 | ) | |
| 113 | warnings.warn(override_message, UserWarning, stacklevel=4) | |
| 114 | else: | |
| 115 | warnings.warn(deprecation_message, FutureWarning, stacklevel=4) | |
| 116 | predicate = op | |
| 117 | if kwargs: | |
| 118 | first = next(iter(kwargs.keys())) | |
| 119 | raise TypeError(f"sjoin() got an unexpected keyword argument '{first}'") | |
| 120 | ||
| 86 | 121 | _basic_checks(left_df, right_df, how, lsuffix, rsuffix) |
| 87 | 122 | |
| 88 | indices = _geom_predicate_query(left_df, right_df, op) | |
| 123 | indices = _geom_predicate_query(left_df, right_df, predicate) | |
| 89 | 124 | |
| 90 | 125 | joined = _frame_join(indices, left_df, right_df, how, lsuffix, rsuffix) |
| 91 | 126 | |
| 142 | 177 | ) |
| 143 | 178 | |
| 144 | 179 | |
| 145 | def _geom_predicate_query(left_df, right_df, op): | |
| 180 | def _geom_predicate_query(left_df, right_df, predicate): | |
| 146 | 181 | """Compute geometric comparisons and get matching indices. |
| 147 | 182 | |
| 148 | 183 | Parameters |
| 149 | 184 | ---------- |
| 150 | 185 | left_df : GeoDataFrame |
| 151 | 186 | right_df : GeoDataFrame |
| 152 | op : string | |
| 187 | predicate : string | |
| 153 | 188 | Binary predicate to query. |
| 154 | 189 | |
| 155 | 190 | Returns |
| 164 | 199 | warnings.filterwarnings( |
| 165 | 200 | "ignore", "Generated spatial index is empty", FutureWarning |
| 166 | 201 | ) |
| 167 | if op == "within": | |
| 202 | ||
| 203 | original_predicate = predicate | |
| 204 | ||
| 205 | if predicate == "within": | |
| 168 | 206 | # within is implemented as the inverse of contains |
| 169 | 207 | # contains is a faster predicate |
| 170 | 208 | # see discussion at https://github.com/geopandas/geopandas/pull/1421 |
| 174 | 212 | else: |
| 175 | 213 | # all other predicates are symmetric |
| 176 | 214 | # keep them the same |
| 177 | predicate = op | |
| 178 | 215 | sindex = right_df.sindex |
| 179 | 216 | input_geoms = left_df.geometry |
| 180 | 217 | |
| 184 | 221 | else: |
| 185 | 222 | # when sindex is empty / has no valid geometries |
| 186 | 223 | indices = pd.DataFrame(columns=["_key_left", "_key_right"], dtype=float) |
| 187 | if op == "within": | |
| 224 | ||
| 225 | if original_predicate == "within": | |
| 188 | 226 | # within is implemented as the inverse of contains |
| 189 | 227 | # flip back the results |
| 190 | 228 | indices = indices.rename( |
| 194 | 232 | return indices |
| 195 | 233 | |
| 196 | 234 | |
| 197 | def _frame_join(indices, left_df, right_df, how, lsuffix, rsuffix): | |
| 235 | def _frame_join(join_df, left_df, right_df, how, lsuffix, rsuffix): | |
| 198 | 236 | """Join the GeoDataFrames at the DataFrame level. |
| 199 | 237 | |
| 200 | 238 | Parameters |
| 201 | 239 | ---------- |
| 202 | indices : DataFrame | |
| 203 | Indexes returned by the geometric join. | |
| 240 | join_df : DataFrame | |
| 241 | Indices and join data returned by the geometric join. | |
| 204 | 242 | Must have columns `_key_left` and `_key_right` |
| 205 | 243 | with integer indices representing the matches |
| 206 | 244 | from `left_df` and `right_df` respectively. |
| 245 | Additional columns may be included and will be copied to | |
| 246 | the resultant GeoDataFrame. | |
| 207 | 247 | left_df : GeoDataFrame |
| 208 | 248 | right_df : GeoDataFrame |
| 209 | 249 | lsuffix : string |
| 252 | 292 | |
| 253 | 293 | # perform join on the dataframes |
| 254 | 294 | if how == "inner": |
| 255 | indices = indices.set_index("_key_left") | |
| 295 | join_df = join_df.set_index("_key_left") | |
| 256 | 296 | joined = ( |
| 257 | left_df.merge(indices, left_index=True, right_index=True) | |
| 297 | left_df.merge(join_df, left_index=True, right_index=True) | |
| 258 | 298 | .merge( |
| 259 | 299 | right_df.drop(right_df.geometry.name, axis=1), |
| 260 | 300 | left_on="_key_right", |
| 270 | 310 | joined.index.name = left_index_name |
| 271 | 311 | |
| 272 | 312 | elif how == "left": |
| 273 | indices = indices.set_index("_key_left") | |
| 313 | join_df = join_df.set_index("_key_left") | |
| 274 | 314 | joined = ( |
| 275 | left_df.merge(indices, left_index=True, right_index=True, how="left") | |
| 315 | left_df.merge(join_df, left_index=True, right_index=True, how="left") | |
| 276 | 316 | .merge( |
| 277 | 317 | right_df.drop(right_df.geometry.name, axis=1), |
| 278 | 318 | how="left", |
| 292 | 332 | joined = ( |
| 293 | 333 | left_df.drop(left_df.geometry.name, axis=1) |
| 294 | 334 | .merge( |
| 295 | indices.merge( | |
| 335 | join_df.merge( | |
| 296 | 336 | right_df, left_on="_key_right", right_index=True, how="right" |
| 297 | 337 | ), |
| 298 | 338 | left_index=True, |
| 299 | 339 | right_on="_key_left", |
| 300 | 340 | how="right", |
| 341 | suffixes=("_{}".format(lsuffix), "_{}".format(rsuffix)), | |
| 301 | 342 | ) |
| 302 | 343 | .set_index(index_right) |
| 303 | 344 | .drop(["_key_left", "_key_right"], axis=1) |
| 308 | 349 | joined.index.name = right_index_name |
| 309 | 350 | |
| 310 | 351 | return joined |
| 352 | ||
| 353 | ||
| 354 | def _nearest_query( | |
| 355 | left_df: GeoDataFrame, | |
| 356 | right_df: GeoDataFrame, | |
| 357 | max_distance: float, | |
| 358 | how: str, | |
| 359 | return_distance: bool, | |
| 360 | ): | |
| 361 | if not (compat.PYGEOS_GE_010 and compat.USE_PYGEOS): | |
| 362 | raise NotImplementedError( | |
| 363 | "Currently, only PyGEOS >= 0.10.0 supports `nearest_all`. " | |
| 364 | + compat.INSTALL_PYGEOS_ERROR | |
| 365 | ) | |
| 366 | # use the opposite of the join direction for the index | |
| 367 | use_left_as_sindex = how == "right" | |
| 368 | if use_left_as_sindex: | |
| 369 | sindex = left_df.sindex | |
| 370 | query = right_df.geometry | |
| 371 | else: | |
| 372 | sindex = right_df.sindex | |
| 373 | query = left_df.geometry | |
| 374 | if sindex: | |
| 375 | res = sindex.nearest( | |
| 376 | query, | |
| 377 | return_all=True, | |
| 378 | max_distance=max_distance, | |
| 379 | return_distance=return_distance, | |
| 380 | ) | |
| 381 | if return_distance: | |
| 382 | (input_idx, tree_idx), distances = res | |
| 383 | else: | |
| 384 | (input_idx, tree_idx) = res | |
| 385 | distances = None | |
| 386 | if use_left_as_sindex: | |
| 387 | l_idx, r_idx = tree_idx, input_idx | |
| 388 | sort_order = np.argsort(l_idx, kind="stable") | |
| 389 | l_idx, r_idx = l_idx[sort_order], r_idx[sort_order] | |
| 390 | if distances is not None: | |
| 391 | distances = distances[sort_order] | |
| 392 | else: | |
| 393 | l_idx, r_idx = input_idx, tree_idx | |
| 394 | join_df = pd.DataFrame( | |
| 395 | {"_key_left": l_idx, "_key_right": r_idx, "distances": distances} | |
| 396 | ) | |
| 397 | else: | |
| 398 | # when sindex is empty / has no valid geometries | |
| 399 | join_df = pd.DataFrame( | |
| 400 | columns=["_key_left", "_key_right", "distances"], dtype=float | |
| 401 | ) | |
| 402 | return join_df | |
| 403 | ||
| 404 | ||
| 405 | def sjoin_nearest( | |
| 406 | left_df: GeoDataFrame, | |
| 407 | right_df: GeoDataFrame, | |
| 408 | how: str = "inner", | |
| 409 | max_distance: Optional[float] = None, | |
| 410 | lsuffix: str = "left", | |
| 411 | rsuffix: str = "right", | |
| 412 | distance_col: Optional[str] = None, | |
| 413 | ) -> GeoDataFrame: | |
| 414 | """Spatial join of two GeoDataFrames based on the distance between their geometries. | |
| 415 | ||
| 416 | Results will include multiple output records for a single input record | |
| 417 | where there are multiple equidistant nearest or intersected neighbors. | |
| 418 | ||
| 419 | See the User Guide page | |
| 420 | https://geopandas.readthedocs.io/en/latest/docs/user_guide/mergingdata.html | |
| 421 | for more details. | |
| 422 | ||
| 423 | ||
| 424 | Parameters | |
| 425 | ---------- | |
| 426 | left_df, right_df : GeoDataFrames | |
| 427 | how : string, default 'inner' | |
| 428 | The type of join: | |
| 429 | ||
| 430 | * 'left': use keys from left_df; retain only left_df geometry column | |
| 431 | * 'right': use keys from right_df; retain only right_df geometry column | |
| 432 | * 'inner': use intersection of keys from both dfs; retain only | |
| 433 | left_df geometry column | |
| 434 | max_distance : float, default None | |
| 435 | Maximum distance within which to query for nearest geometry. | |
| 436 | Must be greater than 0. | |
| 437 | The max_distance used to search for nearest items in the tree may have a | |
| 438 | significant impact on performance by reducing the number of input | |
| 439 | geometries that are evaluated for nearest items in the tree. | |
| 440 | lsuffix : string, default 'left' | |
| 441 | Suffix to apply to overlapping column names (left GeoDataFrame). | |
| 442 | rsuffix : string, default 'right' | |
| 443 | Suffix to apply to overlapping column names (right GeoDataFrame). | |
| 444 | distance_col : string, default None | |
| 445 | If set, save the distances computed between matching geometries under a | |
| 446 | column of this name in the joined GeoDataFrame. | |
| 447 | ||
| 448 | Examples | |
| 449 | -------- | |
| 450 | >>> countries = geopandas.read_file(geopandas.datasets.get_\ | |
| 451 | path("naturalearth_lowres")) | |
| 452 | >>> cities = geopandas.read_file(geopandas.datasets.get_path("naturalearth_cities")) | |
| 453 | >>> countries.head(2).name # doctest: +SKIP | |
| 454 | pop_est continent name \ | |
| 455 | iso_a3 gdp_md_est geometry | |
| 456 | 0 920938 Oceania Fiji FJI 8374.0 MULTIPOLY\ | |
| 457 | GON (((180.00000 -16.06713, 180.00000... | |
| 458 | 1 53950935 Africa Tanzania TZA 150600.0 POLYGON (\ | |
| 459 | (33.90371 -0.95000, 34.07262 -1.05982... | |
| 460 | >>> cities.head(2).name # doctest: +SKIP | |
| 461 | name geometry | |
| 462 | 0 Vatican City POINT (12.45339 41.90328) | |
| 463 | 1 San Marino POINT (12.44177 43.93610) | |
| 464 | ||
| 465 | >>> cities_w_country_data = geopandas.sjoin_nearest(cities, countries) | |
| 466 | >>> cities_w_country_data[['name_left', 'name_right']].head(2) # doctest: +SKIP | |
| 467 | name_left geometry index_right pop_est continent name_\ | |
| 468 | right iso_a3 gdp_md_est | |
| 469 | 0 Vatican City POINT (12.45339 41.90328) 141 62137802 Europe \ | |
| 470 | Italy ITA 2221000.0 | |
| 471 | 1 San Marino POINT (12.44177 43.93610) 141 62137802 Europe \ | |
| 472 | Italy ITA 2221000.0 | |
| 473 | ||
| 474 | To include the distances: | |
| 475 | ||
| 476 | >>> cities_w_country_data = geopandas.sjoin_nearest\ | |
| 477 | (cities, countries, distance_col="distances") | |
| 478 | >>> cities_w_country_data[["name_left", "name_right", \ | |
| 479 | "distances"]].head(2) # doctest: +SKIP | |
| 480 | name_left name_right distances | |
| 481 | 0 Vatican City Italy 0.0 | |
| 482 | 1 San Marino Italy 0.0 | |
| 483 | ||
| 484 | In the following example, we get multiple cities for Italy because all results are | |
| 485 | equidistant (in this case zero because they intersect). | |
| 486 | In fact, we get 3 results in total: | |
| 487 | ||
| 488 | >>> countries_w_city_data = geopandas.sjoin_nearest\ | |
| 489 | (cities, countries, distance_col="distances", how="right") | |
| 490 | >>> italy_results = \ | |
| 491 | countries_w_city_data[countries_w_city_data["name_left"] == "Italy"] | |
| 492 | >>> italy_results # doctest: +SKIP | |
| 493 | name_x name_y | |
| 494 | 141 Vatican City Italy | |
| 495 | 141 San Marino Italy | |
| 496 | 141 Rome Italy | |
| 497 | ||
| 498 | See also | |
| 499 | -------- | |
| 500 | sjoin : binary predicate joins | |
| 501 | GeoDataFrame.sjoin_nearest : equivalent method | |
| 502 | ||
| 503 | Notes | |
| 504 | ----- | |
| 505 | Since this join relies on distances, results will be innaccurate | |
| 506 | if your geometries are in a geographic CRS. | |
| 507 | ||
| 508 | Every operation in GeoPandas is planar, i.e. the potential third | |
| 509 | dimension is not taken into account. | |
| 510 | """ | |
| 511 | _basic_checks(left_df, right_df, how, lsuffix, rsuffix) | |
| 512 | ||
| 513 | left_df.geometry.values.check_geographic_crs(stacklevel=1) | |
| 514 | right_df.geometry.values.check_geographic_crs(stacklevel=1) | |
| 515 | ||
| 516 | return_distance = distance_col is not None | |
| 517 | ||
| 518 | join_df = _nearest_query(left_df, right_df, max_distance, how, return_distance) | |
| 519 | ||
| 520 | if return_distance: | |
| 521 | join_df = join_df.rename(columns={"distances": distance_col}) | |
| 522 | else: | |
| 523 | join_df.pop("distances") | |
| 524 | ||
| 525 | joined = _frame_join(join_df, left_df, right_df, how, lsuffix, rsuffix) | |
| 526 | ||
| 527 | if return_distance: | |
| 528 | columns = [c for c in joined.columns if c != distance_col] + [distance_col] | |
| 529 | joined = joined[columns] | |
| 530 | ||
| 531 | return joined | |
| 0 | 0 | """Tests for the clip module.""" |
| 1 | 1 | |
| 2 | 2 | import warnings |
| 3 | from distutils.version import LooseVersion | |
| 3 | 4 | |
| 4 | 5 | import numpy as np |
| 6 | import pandas as pd | |
| 5 | 7 | |
| 6 | 8 | import shapely |
| 7 | from shapely.geometry import Polygon, Point, LineString, LinearRing, GeometryCollection | |
| 9 | from shapely.geometry import ( | |
| 10 | Polygon, | |
| 11 | Point, | |
| 12 | LineString, | |
| 13 | LinearRing, | |
| 14 | GeometryCollection, | |
| 15 | MultiPoint, | |
| 16 | ) | |
| 8 | 17 | |
| 9 | 18 | import geopandas |
| 10 | 19 | from geopandas import GeoDataFrame, GeoSeries, clip |
| 14 | 23 | |
| 15 | 24 | |
| 16 | 25 | pytestmark = pytest.mark.skip_no_sindex |
| 26 | pandas_133 = pd.__version__ == LooseVersion("1.3.3") | |
| 17 | 27 | |
| 18 | 28 | |
| 19 | 29 | @pytest.fixture |
| 20 | 30 | def point_gdf(): |
| 21 | 31 | """Create a point GeoDataFrame.""" |
| 22 | 32 | pts = np.array([[2, 2], [3, 4], [9, 8], [-12, -15]]) |
| 23 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:4326") | |
| 33 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:3857") | |
| 24 | 34 | return gdf |
| 25 | 35 | |
| 26 | 36 | |
| 29 | 39 | """Create a point GeoDataFrame. Its points are all outside the single |
| 30 | 40 | rectangle, and its bounds are outside the single rectangle's.""" |
| 31 | 41 | pts = np.array([[5, 15], [15, 15], [15, 20]]) |
| 32 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:4326") | |
| 42 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:3857") | |
| 33 | 43 | return gdf |
| 34 | 44 | |
| 35 | 45 | |
| 38 | 48 | """Create a point GeoDataFrame. Its points are all outside the single |
| 39 | 49 | rectangle, and its bounds are overlapping the single rectangle's.""" |
| 40 | 50 | pts = np.array([[5, 15], [15, 15], [15, 5]]) |
| 41 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:4326") | |
| 51 | gdf = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:3857") | |
| 42 | 52 | return gdf |
| 43 | 53 | |
| 44 | 54 | |
| 46 | 56 | def single_rectangle_gdf(): |
| 47 | 57 | """Create a single rectangle for clipping.""" |
| 48 | 58 | poly_inters = Polygon([(0, 0), (0, 10), (10, 10), (10, 0), (0, 0)]) |
| 49 | gdf = GeoDataFrame([1], geometry=[poly_inters], crs="EPSG:4326") | |
| 59 | gdf = GeoDataFrame([1], geometry=[poly_inters], crs="EPSG:3857") | |
| 50 | 60 | gdf["attr2"] = "site-boundary" |
| 51 | 61 | return gdf |
| 52 | 62 | |
| 59 | 69 | eliminates the slivers in the clip return. |
| 60 | 70 | """ |
| 61 | 71 | poly_inters = Polygon([(-5, -5), (-5, 15), (15, 15), (15, -5), (-5, -5)]) |
| 62 | gdf = GeoDataFrame([1], geometry=[poly_inters], crs="EPSG:4326") | |
| 72 | gdf = GeoDataFrame([1], geometry=[poly_inters], crs="EPSG:3857") | |
| 63 | 73 | gdf["attr2"] = ["study area"] |
| 64 | 74 | return gdf |
| 65 | 75 | |
| 87 | 97 | """Create Line Objects For Testing""" |
| 88 | 98 | linea = LineString([(1, 1), (2, 2), (3, 2), (5, 3)]) |
| 89 | 99 | lineb = LineString([(3, 4), (5, 7), (12, 2), (10, 5), (9, 7.5)]) |
| 90 | gdf = GeoDataFrame([1, 2], geometry=[linea, lineb], crs="EPSG:4326") | |
| 100 | gdf = GeoDataFrame([1, 2], geometry=[linea, lineb], crs="EPSG:3857") | |
| 91 | 101 | return gdf |
| 92 | 102 | |
| 93 | 103 | |
| 95 | 105 | def multi_poly_gdf(donut_geometry): |
| 96 | 106 | """Create a multi-polygon GeoDataFrame.""" |
| 97 | 107 | multi_poly = donut_geometry.unary_union |
| 98 | out_df = GeoDataFrame(geometry=GeoSeries(multi_poly), crs="EPSG:4326") | |
| 108 | out_df = GeoDataFrame(geometry=GeoSeries(multi_poly), crs="EPSG:3857") | |
| 99 | 109 | out_df["attr"] = ["pool"] |
| 100 | 110 | return out_df |
| 101 | 111 | |
| 107 | 117 | # Create a single and multi line object |
| 108 | 118 | multiline_feat = two_line_gdf.unary_union |
| 109 | 119 | linec = LineString([(2, 1), (3, 1), (4, 1), (5, 2)]) |
| 110 | out_df = GeoDataFrame(geometry=GeoSeries([multiline_feat, linec]), crs="EPSG:4326") | |
| 120 | out_df = GeoDataFrame(geometry=GeoSeries([multiline_feat, linec]), crs="EPSG:3857") | |
| 111 | 121 | out_df["attr"] = ["road", "stream"] |
| 112 | 122 | return out_df |
| 113 | 123 | |
| 120 | 130 | geometry=GeoSeries( |
| 121 | 131 | [multi_point, Point(2, 5), Point(-11, -14), Point(-10, -12)] |
| 122 | 132 | ), |
| 123 | crs="EPSG:4326", | |
| 133 | crs="EPSG:3857", | |
| 124 | 134 | ) |
| 125 | 135 | out_df["attr"] = ["tree", "another tree", "shrub", "berries"] |
| 126 | 136 | return out_df |
| 134 | 144 | poly = Polygon([(3, 4), (5, 2), (12, 2), (10, 5), (9, 7.5)]) |
| 135 | 145 | ring = LinearRing([(1, 1), (2, 2), (3, 2), (5, 3), (12, 1)]) |
| 136 | 146 | gdf = GeoDataFrame( |
| 137 | [1, 2, 3, 4], geometry=[point, poly, line, ring], crs="EPSG:4326" | |
| 147 | [1, 2, 3, 4], geometry=[point, poly, line, ring], crs="EPSG:3857" | |
| 138 | 148 | ) |
| 139 | 149 | return gdf |
| 140 | 150 | |
| 145 | 155 | point = Point([(2, 3), (11, 4), (7, 2), (8, 9), (1, 13)]) |
| 146 | 156 | poly = Polygon([(3, 4), (5, 2), (12, 2), (10, 5), (9, 7.5)]) |
| 147 | 157 | coll = GeometryCollection([point, poly]) |
| 148 | gdf = GeoDataFrame([1], geometry=[coll], crs="EPSG:4326") | |
| 158 | gdf = GeoDataFrame([1], geometry=[coll], crs="EPSG:3857") | |
| 149 | 159 | return gdf |
| 150 | 160 | |
| 151 | 161 | |
| 154 | 164 | """Create a line that will create a point when clipped.""" |
| 155 | 165 | linea = LineString([(10, 5), (13, 5), (15, 5)]) |
| 156 | 166 | lineb = LineString([(1, 1), (2, 2), (3, 2), (5, 3), (12, 1)]) |
| 157 | gdf = GeoDataFrame([1, 2], geometry=[linea, lineb], crs="EPSG:4326") | |
| 167 | gdf = GeoDataFrame([1, 2], geometry=[linea, lineb], crs="EPSG:3857") | |
| 158 | 168 | return gdf |
| 159 | 169 | |
| 160 | 170 | |
| 181 | 191 | def test_non_overlapping_geoms(): |
| 182 | 192 | """Test that a bounding box returns empty if the extents don't overlap""" |
| 183 | 193 | unit_box = Polygon([(0, 0), (0, 1), (1, 1), (1, 0), (0, 0)]) |
| 184 | unit_gdf = GeoDataFrame([1], geometry=[unit_box], crs="EPSG:4326") | |
| 194 | unit_gdf = GeoDataFrame([1], geometry=[unit_box], crs="EPSG:3857") | |
| 185 | 195 | non_overlapping_gdf = unit_gdf.copy() |
| 186 | 196 | non_overlapping_gdf = non_overlapping_gdf.geometry.apply( |
| 187 | 197 | lambda x: shapely.affinity.translate(x, xoff=20) |
| 196 | 206 | """Test clipping a points GDF with a generic polygon geometry.""" |
| 197 | 207 | clip_pts = clip(point_gdf, single_rectangle_gdf) |
| 198 | 208 | pts = np.array([[2, 2], [3, 4], [9, 8]]) |
| 199 | exp = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:4326") | |
| 209 | exp = GeoDataFrame([Point(xy) for xy in pts], columns=["geometry"], crs="EPSG:3857") | |
| 200 | 210 | assert_geodataframe_equal(clip_pts, exp) |
| 201 | 211 | |
| 202 | 212 | |
| 208 | 218 | exp = GeoDataFrame( |
| 209 | 219 | [Point(xy) for xy in pts], |
| 210 | 220 | columns=["geometry2"], |
| 211 | crs="EPSG:4326", | |
| 221 | crs="EPSG:3857", | |
| 212 | 222 | geometry="geometry2", |
| 213 | 223 | ) |
| 214 | 224 | assert_geodataframe_equal(clip_pts, exp) |
| 238 | 248 | assert all(clipped_poly.geom_type == "Polygon") |
| 239 | 249 | |
| 240 | 250 | |
| 251 | @pytest.mark.xfail(pandas_133, reason="Regression in pandas 1.3.3 (GH #2101)") | |
| 241 | 252 | def test_clip_multipoly_keep_slivers(multi_poly_gdf, single_rectangle_gdf): |
| 242 | 253 | """Test a multi poly object where the return includes a sliver. |
| 243 | 254 | Also the bounds of the object should == the bounds of the clip object |
| 248 | 259 | assert "GeometryCollection" in clipped.geom_type[0] |
| 249 | 260 | |
| 250 | 261 | |
| 262 | @pytest.mark.xfail(pandas_133, reason="Regression in pandas 1.3.3 (GH #2101)") | |
| 251 | 263 | def test_clip_multipoly_keep_geom_type(multi_poly_gdf, single_rectangle_gdf): |
| 252 | 264 | """Test a multi poly object where the return includes a sliver. |
| 253 | 265 | Also the bounds of the object should == the bounds of the clip object |
| 281 | 293 | assert clipped.geom_type[0] == "MultiPoint" |
| 282 | 294 | assert hasattr(clipped, "attr") |
| 283 | 295 | # All points should intersect the clip geom |
| 296 | assert len(clipped) == 2 | |
| 297 | clipped_mutltipoint = MultiPoint( | |
| 298 | [ | |
| 299 | Point(2, 2), | |
| 300 | Point(3, 4), | |
| 301 | Point(9, 8), | |
| 302 | ] | |
| 303 | ) | |
| 304 | assert clipped.iloc[0].geometry.wkt == clipped_mutltipoint.wkt | |
| 284 | 305 | assert all(clipped.intersects(single_rectangle_gdf.unary_union)) |
| 285 | 306 | |
| 286 | 307 | |
| 334 | 355 | exp_poly = polygon.intersection( |
| 335 | 356 | Polygon([(0, 0), (0, 10), (10, 10), (10, 0), (0, 0)]) |
| 336 | 357 | ) |
| 337 | exp = GeoDataFrame([1], geometry=[exp_poly], crs="EPSG:4326") | |
| 358 | exp = GeoDataFrame([1], geometry=[exp_poly], crs="EPSG:3857") | |
| 338 | 359 | exp["attr2"] = "site-boundary" |
| 339 | 360 | assert_geodataframe_equal(clipped, exp) |
| 340 | 361 | |
| 364 | 385 | |
| 365 | 386 | |
| 366 | 387 | def test_clip_box_overlap(pointsoutside_overlap_gdf, single_rectangle_gdf): |
| 367 | """Test clip when intersection is emtpy and boxes do overlap.""" | |
| 388 | """Test clip when intersection is empty and boxes do overlap.""" | |
| 368 | 389 | clipped = clip(pointsoutside_overlap_gdf, single_rectangle_gdf) |
| 369 | 390 | assert len(clipped) == 0 |
| 370 | 391 | |
| 385 | 406 | |
| 386 | 407 | def test_warning_crs_mismatch(point_gdf, single_rectangle_gdf): |
| 387 | 408 | with pytest.warns(UserWarning, match="CRS mismatch between the CRS"): |
| 388 | clip(point_gdf, single_rectangle_gdf.to_crs(3857)) | |
| 409 | clip(point_gdf, single_rectangle_gdf.to_crs(4326)) | |
| 0 | 0 | from distutils.version import LooseVersion |
| 1 | import math | |
| 2 | from typing import Sequence | |
| 3 | from geopandas.testing import assert_geodataframe_equal | |
| 1 | 4 | |
| 2 | 5 | import numpy as np |
| 3 | 6 | import pandas as pd |
| 5 | 8 | from shapely.geometry import Point, Polygon, GeometryCollection |
| 6 | 9 | |
| 7 | 10 | import geopandas |
| 8 | from geopandas import GeoDataFrame, GeoSeries, read_file, sjoin | |
| 11 | import geopandas._compat as compat | |
| 12 | from geopandas import GeoDataFrame, GeoSeries, read_file, sjoin, sjoin_nearest | |
| 13 | from geopandas.testing import assert_geoseries_equal | |
| 9 | 14 | |
| 10 | 15 | from pandas.testing import assert_frame_equal |
| 11 | 16 | import pytest |
| 17 | ||
| 18 | ||
| 19 | TEST_NEAREST = compat.PYGEOS_GE_010 and compat.USE_PYGEOS | |
| 12 | 20 | |
| 13 | 21 | |
| 14 | 22 | pytestmark = pytest.mark.skip_no_sindex |
| 88 | 96 | |
| 89 | 97 | |
| 90 | 98 | class TestSpatialJoin: |
| 99 | @pytest.mark.parametrize( | |
| 100 | "how, lsuffix, rsuffix, expected_cols", | |
| 101 | [ | |
| 102 | ("left", "left", "right", {"col_left", "col_right", "index_right"}), | |
| 103 | ("inner", "left", "right", {"col_left", "col_right", "index_right"}), | |
| 104 | ("right", "left", "right", {"col_left", "col_right", "index_left"}), | |
| 105 | ("left", "lft", "rgt", {"col_lft", "col_rgt", "index_rgt"}), | |
| 106 | ("inner", "lft", "rgt", {"col_lft", "col_rgt", "index_rgt"}), | |
| 107 | ("right", "lft", "rgt", {"col_lft", "col_rgt", "index_lft"}), | |
| 108 | ], | |
| 109 | ) | |
| 110 | def test_suffixes(self, how: str, lsuffix: str, rsuffix: str, expected_cols): | |
| 111 | left = GeoDataFrame({"col": [1], "geometry": [Point(0, 0)]}) | |
| 112 | right = GeoDataFrame({"col": [1], "geometry": [Point(0, 0)]}) | |
| 113 | joined = sjoin(left, right, how=how, lsuffix=lsuffix, rsuffix=rsuffix) | |
| 114 | assert set(joined.columns) == expected_cols | set(("geometry",)) | |
| 115 | ||
| 91 | 116 | @pytest.mark.parametrize("dfs", ["default-index", "string-index"], indirect=True) |
| 92 | 117 | def test_crs_mismatch(self, dfs): |
| 93 | 118 | index, df1, df2, expected = dfs |
| 95 | 120 | with pytest.warns(UserWarning, match="CRS mismatch between the CRS"): |
| 96 | 121 | sjoin(df1, df2) |
| 97 | 122 | |
| 123 | @pytest.mark.parametrize("dfs", ["default-index"], indirect=True) | |
| 124 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 125 | def test_deprecated_op_param(self, dfs, op): | |
| 126 | _, df1, df2, _ = dfs | |
| 127 | with pytest.warns(FutureWarning, match="`op` parameter is deprecated"): | |
| 128 | sjoin(df1, df2, op=op) | |
| 129 | ||
| 130 | @pytest.mark.parametrize("dfs", ["default-index"], indirect=True) | |
| 131 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 132 | @pytest.mark.parametrize("predicate", ["contains", "within"]) | |
| 133 | def test_deprecated_op_param_nondefault_predicate(self, dfs, op, predicate): | |
| 134 | _, df1, df2, _ = dfs | |
| 135 | match = "use the `predicate` parameter instead" | |
| 136 | if op != predicate: | |
| 137 | warntype = UserWarning | |
| 138 | match = ( | |
| 139 | "`predicate` will be overriden by the value of `op`" | |
| 140 | + r"(.|\s)*" | |
| 141 | + match | |
| 142 | ) | |
| 143 | else: | |
| 144 | warntype = FutureWarning | |
| 145 | with pytest.warns(warntype, match=match): | |
| 146 | sjoin(df1, df2, predicate=predicate, op=op) | |
| 147 | ||
| 148 | @pytest.mark.parametrize("dfs", ["default-index"], indirect=True) | |
| 149 | def test_unknown_kwargs(self, dfs): | |
| 150 | _, df1, df2, _ = dfs | |
| 151 | with pytest.raises( | |
| 152 | TypeError, | |
| 153 | match=r"sjoin\(\) got an unexpected keyword argument 'extra_param'", | |
| 154 | ): | |
| 155 | sjoin(df1, df2, extra_param="test") | |
| 156 | ||
| 157 | @pytest.mark.filterwarnings("ignore:The `op` parameter:FutureWarning") | |
| 98 | 158 | @pytest.mark.parametrize( |
| 99 | 159 | "dfs", |
| 100 | 160 | [ |
| 106 | 166 | ], |
| 107 | 167 | indirect=True, |
| 108 | 168 | ) |
| 109 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 110 | def test_inner(self, op, dfs): | |
| 169 | @pytest.mark.parametrize("predicate", ["intersects", "contains", "within"]) | |
| 170 | @pytest.mark.parametrize("predicate_kw", ["predicate", "op"]) | |
| 171 | def test_inner(self, predicate, predicate_kw, dfs): | |
| 111 | 172 | index, df1, df2, expected = dfs |
| 112 | 173 | |
| 113 | res = sjoin(df1, df2, how="inner", op=op) | |
| 114 | ||
| 115 | exp = expected[op].dropna().copy() | |
| 174 | res = sjoin(df1, df2, how="inner", **{predicate_kw: predicate}) | |
| 175 | ||
| 176 | exp = expected[predicate].dropna().copy() | |
| 116 | 177 | exp = exp.drop("geometry_y", axis=1).rename(columns={"geometry_x": "geometry"}) |
| 117 | 178 | exp[["df1", "df2"]] = exp[["df1", "df2"]].astype("int64") |
| 118 | 179 | if index == "default-index": |
| 149 | 210 | ], |
| 150 | 211 | indirect=True, |
| 151 | 212 | ) |
| 152 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 153 | def test_left(self, op, dfs): | |
| 213 | @pytest.mark.parametrize("predicate", ["intersects", "contains", "within"]) | |
| 214 | def test_left(self, predicate, dfs): | |
| 154 | 215 | index, df1, df2, expected = dfs |
| 155 | 216 | |
| 156 | res = sjoin(df1, df2, how="left", op=op) | |
| 217 | res = sjoin(df1, df2, how="left", predicate=predicate) | |
| 157 | 218 | |
| 158 | 219 | if index in ["default-index", "string-index"]: |
| 159 | exp = expected[op].dropna(subset=["index_left"]).copy() | |
| 220 | exp = expected[predicate].dropna(subset=["index_left"]).copy() | |
| 160 | 221 | elif index == "named-index": |
| 161 | exp = expected[op].dropna(subset=["df1_ix"]).copy() | |
| 222 | exp = expected[predicate].dropna(subset=["df1_ix"]).copy() | |
| 162 | 223 | elif index == "multi-index": |
| 163 | exp = expected[op].dropna(subset=["level_0_x"]).copy() | |
| 224 | exp = expected[predicate].dropna(subset=["level_0_x"]).copy() | |
| 164 | 225 | elif index == "named-multi-index": |
| 165 | exp = expected[op].dropna(subset=["df1_ix1"]).copy() | |
| 226 | exp = expected[predicate].dropna(subset=["df1_ix1"]).copy() | |
| 166 | 227 | exp = exp.drop("geometry_y", axis=1).rename(columns={"geometry_x": "geometry"}) |
| 167 | 228 | exp["df1"] = exp["df1"].astype("int64") |
| 168 | 229 | if index == "default-index": |
| 200 | 261 | } |
| 201 | 262 | ) |
| 202 | 263 | not_in = geopandas.GeoDataFrame({"col1": [1], "geometry": [Point(-0.5, 0.5)]}) |
| 203 | empty = sjoin(not_in, polygons, how="left", op="intersects") | |
| 264 | empty = sjoin(not_in, polygons, how="left", predicate="intersects") | |
| 204 | 265 | assert empty.index_right.isnull().all() |
| 205 | empty = sjoin(not_in, polygons, how="right", op="intersects") | |
| 266 | empty = sjoin(not_in, polygons, how="right", predicate="intersects") | |
| 206 | 267 | assert empty.index_left.isnull().all() |
| 207 | empty = sjoin(not_in, polygons, how="inner", op="intersects") | |
| 268 | empty = sjoin(not_in, polygons, how="inner", predicate="intersects") | |
| 208 | 269 | assert empty.empty |
| 209 | 270 | |
| 210 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 271 | @pytest.mark.parametrize("predicate", ["intersects", "contains", "within"]) | |
| 211 | 272 | @pytest.mark.parametrize( |
| 212 | 273 | "empty", |
| 213 | 274 | [ |
| 215 | 276 | GeoDataFrame(geometry=GeoSeries()), |
| 216 | 277 | ], |
| 217 | 278 | ) |
| 218 | def test_join_with_empty(self, op, empty): | |
| 279 | def test_join_with_empty(self, predicate, empty): | |
| 219 | 280 | # Check joins with empty geometry columns/dataframes. |
| 220 | 281 | polygons = geopandas.GeoDataFrame( |
| 221 | 282 | { |
| 226 | 287 | ], |
| 227 | 288 | } |
| 228 | 289 | ) |
| 229 | result = sjoin(empty, polygons, how="left", op=op) | |
| 290 | result = sjoin(empty, polygons, how="left", predicate=predicate) | |
| 230 | 291 | assert result.index_right.isnull().all() |
| 231 | result = sjoin(empty, polygons, how="right", op=op) | |
| 292 | result = sjoin(empty, polygons, how="right", predicate=predicate) | |
| 232 | 293 | assert result.index_left.isnull().all() |
| 233 | result = sjoin(empty, polygons, how="inner", op=op) | |
| 294 | result = sjoin(empty, polygons, how="inner", predicate=predicate) | |
| 234 | 295 | assert result.empty |
| 235 | 296 | |
| 236 | 297 | @pytest.mark.parametrize("dfs", ["default-index", "string-index"], indirect=True) |
| 254 | 315 | ], |
| 255 | 316 | indirect=True, |
| 256 | 317 | ) |
| 257 | @pytest.mark.parametrize("op", ["intersects", "contains", "within"]) | |
| 258 | def test_right(self, op, dfs): | |
| 318 | @pytest.mark.parametrize("predicate", ["intersects", "contains", "within"]) | |
| 319 | def test_right(self, predicate, dfs): | |
| 259 | 320 | index, df1, df2, expected = dfs |
| 260 | 321 | |
| 261 | res = sjoin(df1, df2, how="right", op=op) | |
| 322 | res = sjoin(df1, df2, how="right", predicate=predicate) | |
| 262 | 323 | |
| 263 | 324 | if index in ["default-index", "string-index"]: |
| 264 | exp = expected[op].dropna(subset=["index_right"]).copy() | |
| 325 | exp = expected[predicate].dropna(subset=["index_right"]).copy() | |
| 265 | 326 | elif index == "named-index": |
| 266 | exp = expected[op].dropna(subset=["df2_ix"]).copy() | |
| 327 | exp = expected[predicate].dropna(subset=["df2_ix"]).copy() | |
| 267 | 328 | elif index == "multi-index": |
| 268 | exp = expected[op].dropna(subset=["level_0_y"]).copy() | |
| 329 | exp = expected[predicate].dropna(subset=["level_0_y"]).copy() | |
| 269 | 330 | elif index == "named-multi-index": |
| 270 | exp = expected[op].dropna(subset=["df2_ix1"]).copy() | |
| 331 | exp = expected[predicate].dropna(subset=["df2_ix1"]).copy() | |
| 271 | 332 | exp = exp.drop("geometry_x", axis=1).rename(columns={"geometry_y": "geometry"}) |
| 272 | 333 | exp["df2"] = exp["df2"].astype("int64") |
| 273 | 334 | if index == "default-index": |
| 292 | 353 | exp.index.names = df2.index.names |
| 293 | 354 | |
| 294 | 355 | # GH 1364 fix of behaviour was done in pandas 1.1.0 |
| 295 | if op == "within" and str(pd.__version__) >= LooseVersion("1.1.0"): | |
| 356 | if predicate == "within" and str(pd.__version__) >= LooseVersion("1.1.0"): | |
| 296 | 357 | exp = exp.sort_index() |
| 297 | 358 | |
| 298 | 359 | assert_frame_equal(res, exp, check_index_type=False) |
| 349 | 410 | df = sjoin(self.pointdf, self.polydf, how="inner") |
| 350 | 411 | assert df.shape == (11, 8) |
| 351 | 412 | |
| 352 | def test_sjoin_op(self): | |
| 413 | def test_sjoin_predicate(self): | |
| 353 | 414 | # points within polygons |
| 354 | df = sjoin(self.pointdf, self.polydf, how="left", op="within") | |
| 415 | df = sjoin(self.pointdf, self.polydf, how="left", predicate="within") | |
| 355 | 416 | assert df.shape == (21, 8) |
| 356 | 417 | assert df.loc[1]["BoroName"] == "Staten Island" |
| 357 | 418 | |
| 358 | 419 | # points contain polygons? never happens so we should have nulls |
| 359 | df = sjoin(self.pointdf, self.polydf, how="left", op="contains") | |
| 420 | df = sjoin(self.pointdf, self.polydf, how="left", predicate="contains") | |
| 360 | 421 | assert df.shape == (21, 8) |
| 361 | 422 | assert np.isnan(df.loc[1]["Shape_Area"]) |
| 362 | 423 | |
| 363 | def test_sjoin_bad_op(self): | |
| 424 | def test_sjoin_bad_predicate(self): | |
| 364 | 425 | # AttributeError: 'Point' object has no attribute 'spandex' |
| 365 | 426 | with pytest.raises(ValueError): |
| 366 | sjoin(self.pointdf, self.polydf, how="left", op="spandex") | |
| 427 | sjoin(self.pointdf, self.polydf, how="left", predicate="spandex") | |
| 367 | 428 | |
| 368 | 429 | def test_sjoin_duplicate_column_name(self): |
| 369 | 430 | pointdf2 = self.pointdf.rename(columns={"pointattr1": "Shape_Area"}) |
| 462 | 523 | df2 = sjoin(self.pointdf, self.polydf.append(empty), how="left") |
| 463 | 524 | assert df2.shape == (21, 8) |
| 464 | 525 | |
| 465 | @pytest.mark.parametrize("op", ["intersects", "within", "contains"]) | |
| 466 | def test_sjoin_no_valid_geoms(self, op): | |
| 526 | @pytest.mark.parametrize("predicate", ["intersects", "within", "contains"]) | |
| 527 | def test_sjoin_no_valid_geoms(self, predicate): | |
| 467 | 528 | """Tests a completely empty GeoDataFrame.""" |
| 468 | 529 | empty = GeoDataFrame(geometry=[], crs=self.pointdf.crs) |
| 469 | assert sjoin(self.pointdf, empty, how="inner", op=op).empty | |
| 470 | assert sjoin(self.pointdf, empty, how="right", op=op).empty | |
| 471 | assert sjoin(empty, self.pointdf, how="inner", op=op).empty | |
| 472 | assert sjoin(empty, self.pointdf, how="left", op=op).empty | |
| 530 | assert sjoin(self.pointdf, empty, how="inner", predicate=predicate).empty | |
| 531 | assert sjoin(self.pointdf, empty, how="right", predicate=predicate).empty | |
| 532 | assert sjoin(empty, self.pointdf, how="inner", predicate=predicate).empty | |
| 533 | assert sjoin(empty, self.pointdf, how="left", predicate=predicate).empty | |
| 534 | ||
| 535 | def test_empty_sjoin_return_duplicated_columns(self): | |
| 536 | ||
| 537 | nybb = geopandas.read_file(geopandas.datasets.get_path("nybb")) | |
| 538 | nybb2 = nybb.copy() | |
| 539 | nybb2.geometry = nybb2.translate(200000) # to get non-overlapping | |
| 540 | ||
| 541 | result = geopandas.sjoin(nybb, nybb2) | |
| 542 | ||
| 543 | assert "BoroCode_right" in result.columns | |
| 544 | assert "BoroCode_left" in result.columns | |
| 473 | 545 | |
| 474 | 546 | |
| 475 | 547 | class TestSpatialJoinNaturalEarth: |
| 484 | 556 | countries = self.world[["geometry", "name"]] |
| 485 | 557 | countries = countries.rename(columns={"name": "country"}) |
| 486 | 558 | cities_with_country = sjoin( |
| 487 | self.cities, countries, how="inner", op="intersects" | |
| 559 | self.cities, countries, how="inner", predicate="intersects" | |
| 488 | 560 | ) |
| 489 | 561 | assert cities_with_country.shape == (172, 4) |
| 562 | ||
| 563 | ||
| 564 | @pytest.mark.skipif( | |
| 565 | TEST_NEAREST, | |
| 566 | reason=("This test can only be run _without_ PyGEOS >= 0.10 installed"), | |
| 567 | ) | |
| 568 | def test_no_nearest_all(): | |
| 569 | df1 = geopandas.GeoDataFrame({"geometry": []}) | |
| 570 | df2 = geopandas.GeoDataFrame({"geometry": []}) | |
| 571 | with pytest.raises( | |
| 572 | NotImplementedError, | |
| 573 | match="Currently, only PyGEOS >= 0.10.0 supports `nearest_all`", | |
| 574 | ): | |
| 575 | sjoin_nearest(df1, df2) | |
| 576 | ||
| 577 | ||
| 578 | @pytest.mark.skipif( | |
| 579 | not TEST_NEAREST, | |
| 580 | reason=( | |
| 581 | "PyGEOS >= 0.10.0" | |
| 582 | " must be installed and activated via the geopandas.compat module to" | |
| 583 | " test sjoin_nearest" | |
| 584 | ), | |
| 585 | ) | |
| 586 | class TestNearest: | |
| 587 | @pytest.mark.parametrize( | |
| 588 | "how_kwargs", ({}, {"how": "inner"}, {"how": "left"}, {"how": "right"}) | |
| 589 | ) | |
| 590 | def test_allowed_hows(self, how_kwargs): | |
| 591 | left = geopandas.GeoDataFrame({"geometry": []}) | |
| 592 | right = geopandas.GeoDataFrame({"geometry": []}) | |
| 593 | sjoin_nearest(left, right, **how_kwargs) # no error | |
| 594 | ||
| 595 | @pytest.mark.parametrize("how", ("outer", "abcde")) | |
| 596 | def test_invalid_hows(self, how: str): | |
| 597 | left = geopandas.GeoDataFrame({"geometry": []}) | |
| 598 | right = geopandas.GeoDataFrame({"geometry": []}) | |
| 599 | with pytest.raises(ValueError, match="`how` was"): | |
| 600 | sjoin_nearest(left, right, how=how) | |
| 601 | ||
| 602 | @pytest.mark.parametrize("distance_col", (None, "distance")) | |
| 603 | def test_empty_right_df_how_left(self, distance_col: str): | |
| 604 | # all records from left and no results from right | |
| 605 | left = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 606 | right = geopandas.GeoDataFrame({"geometry": []}) | |
| 607 | joined = sjoin_nearest( | |
| 608 | left, | |
| 609 | right, | |
| 610 | how="left", | |
| 611 | distance_col=distance_col, | |
| 612 | ) | |
| 613 | assert_geoseries_equal(joined["geometry"], left["geometry"]) | |
| 614 | assert joined["index_right"].isna().all() | |
| 615 | if distance_col is not None: | |
| 616 | assert joined[distance_col].isna().all() | |
| 617 | ||
| 618 | @pytest.mark.parametrize("distance_col", (None, "distance")) | |
| 619 | def test_empty_right_df_how_right(self, distance_col: str): | |
| 620 | # no records in joined | |
| 621 | left = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 622 | right = geopandas.GeoDataFrame({"geometry": []}) | |
| 623 | joined = sjoin_nearest( | |
| 624 | left, | |
| 625 | right, | |
| 626 | how="right", | |
| 627 | distance_col=distance_col, | |
| 628 | ) | |
| 629 | assert joined.empty | |
| 630 | if distance_col is not None: | |
| 631 | assert distance_col in joined | |
| 632 | ||
| 633 | @pytest.mark.parametrize("how", ["inner", "left"]) | |
| 634 | @pytest.mark.parametrize("distance_col", (None, "distance")) | |
| 635 | def test_empty_left_df(self, how, distance_col: str): | |
| 636 | right = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 637 | left = geopandas.GeoDataFrame({"geometry": []}) | |
| 638 | joined = sjoin_nearest(left, right, how=how, distance_col=distance_col) | |
| 639 | assert joined.empty | |
| 640 | if distance_col is not None: | |
| 641 | assert distance_col in joined | |
| 642 | ||
| 643 | @pytest.mark.parametrize("distance_col", (None, "distance")) | |
| 644 | def test_empty_left_df_how_right(self, distance_col: str): | |
| 645 | right = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 646 | left = geopandas.GeoDataFrame({"geometry": []}) | |
| 647 | joined = sjoin_nearest( | |
| 648 | left, | |
| 649 | right, | |
| 650 | how="right", | |
| 651 | distance_col=distance_col, | |
| 652 | ) | |
| 653 | assert_geoseries_equal(joined["geometry"], right["geometry"]) | |
| 654 | assert joined["index_left"].isna().all() | |
| 655 | if distance_col is not None: | |
| 656 | assert joined[distance_col].isna().all() | |
| 657 | ||
| 658 | @pytest.mark.parametrize("how", ["inner", "left"]) | |
| 659 | def test_empty_join_due_to_max_distance(self, how): | |
| 660 | # after applying max_distance the join comes back empty | |
| 661 | # (as in NaN in the joined columns) | |
| 662 | left = geopandas.GeoDataFrame({"geometry": [Point(0, 0)]}) | |
| 663 | right = geopandas.GeoDataFrame({"geometry": [Point(1, 1), Point(2, 2)]}) | |
| 664 | joined = sjoin_nearest( | |
| 665 | left, | |
| 666 | right, | |
| 667 | how=how, | |
| 668 | max_distance=1, | |
| 669 | distance_col="distances", | |
| 670 | ) | |
| 671 | expected = left.copy() | |
| 672 | expected["index_right"] = [np.nan] | |
| 673 | expected["distances"] = [np.nan] | |
| 674 | if how == "inner": | |
| 675 | expected = expected.dropna() | |
| 676 | expected["index_right"] = expected["index_right"].astype("int64") | |
| 677 | assert_geodataframe_equal(joined, expected) | |
| 678 | ||
| 679 | def test_empty_join_due_to_max_distance_how_right(self): | |
| 680 | # after applying max_distance the join comes back empty | |
| 681 | # (as in NaN in the joined columns) | |
| 682 | left = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 683 | right = geopandas.GeoDataFrame({"geometry": [Point(2, 2)]}) | |
| 684 | joined = sjoin_nearest( | |
| 685 | left, | |
| 686 | right, | |
| 687 | how="right", | |
| 688 | max_distance=1, | |
| 689 | distance_col="distances", | |
| 690 | ) | |
| 691 | expected = right.copy() | |
| 692 | expected["index_left"] = [np.nan] | |
| 693 | expected["distances"] = [np.nan] | |
| 694 | expected = expected[["index_left", "geometry", "distances"]] | |
| 695 | assert_geodataframe_equal(joined, expected) | |
| 696 | ||
| 697 | @pytest.mark.parametrize("how", ["inner", "left"]) | |
| 698 | def test_max_distance(self, how): | |
| 699 | left = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 700 | right = geopandas.GeoDataFrame({"geometry": [Point(1, 1), Point(2, 2)]}) | |
| 701 | joined = sjoin_nearest( | |
| 702 | left, | |
| 703 | right, | |
| 704 | how=how, | |
| 705 | max_distance=1, | |
| 706 | distance_col="distances", | |
| 707 | ) | |
| 708 | expected = left.copy() | |
| 709 | expected["index_right"] = [np.nan, 0] | |
| 710 | expected["distances"] = [np.nan, 0] | |
| 711 | if how == "inner": | |
| 712 | expected = expected.dropna() | |
| 713 | expected["index_right"] = expected["index_right"].astype("int64") | |
| 714 | assert_geodataframe_equal(joined, expected) | |
| 715 | ||
| 716 | def test_max_distance_how_right(self): | |
| 717 | left = geopandas.GeoDataFrame({"geometry": [Point(1, 1), Point(2, 2)]}) | |
| 718 | right = geopandas.GeoDataFrame({"geometry": [Point(0, 0), Point(1, 1)]}) | |
| 719 | joined = sjoin_nearest( | |
| 720 | left, | |
| 721 | right, | |
| 722 | how="right", | |
| 723 | max_distance=1, | |
| 724 | distance_col="distances", | |
| 725 | ) | |
| 726 | expected = right.copy() | |
| 727 | expected["index_left"] = [np.nan, 0] | |
| 728 | expected["distances"] = [np.nan, 0] | |
| 729 | expected = expected[["index_left", "geometry", "distances"]] | |
| 730 | assert_geodataframe_equal(joined, expected) | |
| 731 | ||
| 732 | @pytest.mark.parametrize("how", ["inner", "left"]) | |
| 733 | @pytest.mark.parametrize( | |
| 734 | "geo_left, geo_right, expected_left, expected_right, distances", | |
| 735 | [ | |
| 736 | ( | |
| 737 | [Point(0, 0), Point(1, 1)], | |
| 738 | [Point(1, 1)], | |
| 739 | [0, 1], | |
| 740 | [0, 0], | |
| 741 | [math.sqrt(2), 0], | |
| 742 | ), | |
| 743 | ( | |
| 744 | [Point(0, 0), Point(1, 1)], | |
| 745 | [Point(1, 1), Point(0, 0)], | |
| 746 | [0, 1], | |
| 747 | [1, 0], | |
| 748 | [0, 0], | |
| 749 | ), | |
| 750 | ( | |
| 751 | [Point(0, 0), Point(1, 1)], | |
| 752 | [Point(1, 1), Point(0, 0), Point(0, 0)], | |
| 753 | [0, 0, 1], | |
| 754 | [1, 2, 0], | |
| 755 | [0, 0, 0], | |
| 756 | ), | |
| 757 | ( | |
| 758 | [Point(0, 0), Point(1, 1)], | |
| 759 | [Point(1, 1), Point(0, 0), Point(2, 2)], | |
| 760 | [0, 1], | |
| 761 | [1, 0], | |
| 762 | [0, 0], | |
| 763 | ), | |
| 764 | ( | |
| 765 | [Point(0, 0), Point(1, 1)], | |
| 766 | [Point(1, 1), Point(0.25, 1)], | |
| 767 | [0, 1], | |
| 768 | [1, 0], | |
| 769 | [math.sqrt(0.25 ** 2 + 1), 0], | |
| 770 | ), | |
| 771 | ( | |
| 772 | [Point(0, 0), Point(1, 1)], | |
| 773 | [Point(-10, -10), Point(100, 100)], | |
| 774 | [0, 1], | |
| 775 | [0, 0], | |
| 776 | [math.sqrt(10 ** 2 + 10 ** 2), math.sqrt(11 ** 2 + 11 ** 2)], | |
| 777 | ), | |
| 778 | ( | |
| 779 | [Point(0, 0), Point(1, 1)], | |
| 780 | [Point(x, y) for x, y in zip(np.arange(10), np.arange(10))], | |
| 781 | [0, 1], | |
| 782 | [0, 1], | |
| 783 | [0, 0], | |
| 784 | ), | |
| 785 | ( | |
| 786 | [Point(0, 0), Point(1, 1), Point(0, 0)], | |
| 787 | [Point(1.1, 1.1), Point(0, 0)], | |
| 788 | [0, 1, 2], | |
| 789 | [1, 0, 1], | |
| 790 | [0, np.sqrt(0.1 ** 2 + 0.1 ** 2), 0], | |
| 791 | ), | |
| 792 | ], | |
| 793 | ) | |
| 794 | def test_sjoin_nearest_left( | |
| 795 | self, | |
| 796 | geo_left, | |
| 797 | geo_right, | |
| 798 | expected_left: Sequence[int], | |
| 799 | expected_right: Sequence[int], | |
| 800 | distances: Sequence[float], | |
| 801 | how, | |
| 802 | ): | |
| 803 | left = geopandas.GeoDataFrame({"geometry": geo_left}) | |
| 804 | right = geopandas.GeoDataFrame({"geometry": geo_right}) | |
| 805 | expected_gdf = left.iloc[expected_left].copy() | |
| 806 | expected_gdf["index_right"] = expected_right | |
| 807 | # without distance col | |
| 808 | joined = sjoin_nearest(left, right, how=how) | |
| 809 | # inner / left join give a different row order | |
| 810 | check_like = how == "inner" | |
| 811 | assert_geodataframe_equal(expected_gdf, joined, check_like=check_like) | |
| 812 | # with distance col | |
| 813 | expected_gdf["distance_col"] = np.array(distances, dtype=float) | |
| 814 | joined = sjoin_nearest(left, right, how=how, distance_col="distance_col") | |
| 815 | assert_geodataframe_equal(expected_gdf, joined, check_like=check_like) | |
| 816 | ||
| 817 | @pytest.mark.parametrize( | |
| 818 | "geo_left, geo_right, expected_left, expected_right, distances", | |
| 819 | [ | |
| 820 | ([Point(0, 0), Point(1, 1)], [Point(1, 1)], [1], [0], [0]), | |
| 821 | ( | |
| 822 | [Point(0, 0), Point(1, 1)], | |
| 823 | [Point(1, 1), Point(0, 0)], | |
| 824 | [1, 0], | |
| 825 | [0, 1], | |
| 826 | [0, 0], | |
| 827 | ), | |
| 828 | ( | |
| 829 | [Point(0, 0), Point(1, 1)], | |
| 830 | [Point(1, 1), Point(0, 0), Point(0, 0)], | |
| 831 | [1, 0, 0], | |
| 832 | [0, 1, 2], | |
| 833 | [0, 0, 0], | |
| 834 | ), | |
| 835 | ( | |
| 836 | [Point(0, 0), Point(1, 1)], | |
| 837 | [Point(1, 1), Point(0, 0), Point(2, 2)], | |
| 838 | [1, 0, 1], | |
| 839 | [0, 1, 2], | |
| 840 | [0, 0, math.sqrt(2)], | |
| 841 | ), | |
| 842 | ( | |
| 843 | [Point(0, 0), Point(1, 1)], | |
| 844 | [Point(1, 1), Point(0.25, 1)], | |
| 845 | [1, 1], | |
| 846 | [0, 1], | |
| 847 | [0, 0.75], | |
| 848 | ), | |
| 849 | ( | |
| 850 | [Point(0, 0), Point(1, 1)], | |
| 851 | [Point(-10, -10), Point(100, 100)], | |
| 852 | [0, 1], | |
| 853 | [0, 1], | |
| 854 | [math.sqrt(10 ** 2 + 10 ** 2), math.sqrt(99 ** 2 + 99 ** 2)], | |
| 855 | ), | |
| 856 | ( | |
| 857 | [Point(0, 0), Point(1, 1)], | |
| 858 | [Point(x, y) for x, y in zip(np.arange(10), np.arange(10))], | |
| 859 | [0, 1] + [1] * 8, | |
| 860 | list(range(10)), | |
| 861 | [0, 0] + [np.sqrt(x ** 2 + x ** 2) for x in np.arange(1, 9)], | |
| 862 | ), | |
| 863 | ( | |
| 864 | [Point(0, 0), Point(1, 1), Point(0, 0)], | |
| 865 | [Point(1.1, 1.1), Point(0, 0)], | |
| 866 | [1, 0, 2], | |
| 867 | [0, 1, 1], | |
| 868 | [np.sqrt(0.1 ** 2 + 0.1 ** 2), 0, 0], | |
| 869 | ), | |
| 870 | ], | |
| 871 | ) | |
| 872 | def test_sjoin_nearest_right( | |
| 873 | self, | |
| 874 | geo_left, | |
| 875 | geo_right, | |
| 876 | expected_left: Sequence[int], | |
| 877 | expected_right: Sequence[int], | |
| 878 | distances: Sequence[float], | |
| 879 | ): | |
| 880 | left = geopandas.GeoDataFrame({"geometry": geo_left}) | |
| 881 | right = geopandas.GeoDataFrame({"geometry": geo_right}) | |
| 882 | expected_gdf = right.iloc[expected_right].copy() | |
| 883 | expected_gdf["index_left"] = expected_left | |
| 884 | expected_gdf = expected_gdf[["index_left", "geometry"]] | |
| 885 | # without distance col | |
| 886 | joined = sjoin_nearest(left, right, how="right") | |
| 887 | assert_geodataframe_equal(expected_gdf, joined) | |
| 888 | # with distance col | |
| 889 | expected_gdf["distance_col"] = np.array(distances, dtype=float) | |
| 890 | joined = sjoin_nearest(left, right, how="right", distance_col="distance_col") | |
| 891 | assert_geodataframe_equal(expected_gdf, joined) | |
| 892 | ||
| 893 | @pytest.mark.filterwarnings("ignore:Geometry is in a geographic CRS") | |
| 894 | def test_sjoin_nearest_inner(self): | |
| 895 | # check equivalency of left and inner join | |
| 896 | countries = read_file(geopandas.datasets.get_path("naturalearth_lowres")) | |
| 897 | cities = read_file(geopandas.datasets.get_path("naturalearth_cities")) | |
| 898 | countries = countries[["geometry", "name"]].rename(columns={"name": "country"}) | |
| 899 | ||
| 900 | # default: inner and left give the same result | |
| 901 | result1 = sjoin_nearest(cities, countries, distance_col="dist") | |
| 902 | assert result1.shape[0] == cities.shape[0] | |
| 903 | result2 = sjoin_nearest(cities, countries, distance_col="dist", how="inner") | |
| 904 | assert_geodataframe_equal(result2, result1) | |
| 905 | result3 = sjoin_nearest(cities, countries, distance_col="dist", how="left") | |
| 906 | assert_geodataframe_equal(result3, result1, check_like=True) | |
| 907 | ||
| 908 | # with max_distance: rows that go above are dropped in case of inner | |
| 909 | result4 = sjoin_nearest(cities, countries, distance_col="dist", max_distance=1) | |
| 910 | assert_geodataframe_equal( | |
| 911 | result4, result1[result1["dist"] < 1], check_like=True | |
| 912 | ) | |
| 913 | result5 = sjoin_nearest( | |
| 914 | cities, countries, distance_col="dist", max_distance=1, how="left" | |
| 915 | ) | |
| 916 | assert result5.shape[0] == cities.shape[0] | |
| 917 | result5 = result5.dropna() | |
| 918 | result5["index_right"] = result5["index_right"].astype("int64") | |
| 919 | assert_geodataframe_equal(result5, result4, check_like=True) | |
| 33 | 33 | # Point and MultiPoint... or even just MultiPoint |
| 34 | 34 | t = x[0].type |
| 35 | 35 | if not all(g.type == t for g in x): |
| 36 | raise ValueError("Geometry type must be homogenous") | |
| 36 | raise ValueError("Geometry type must be homogeneous") | |
| 37 | 37 | if len(x) > 1 and t.startswith("Multi"): |
| 38 | 38 | raise ValueError("Cannot collect {0}. Must have single geometries".format(t)) |
| 39 | 39 |
| 0 | 0 | # required |
| 1 | 1 | fiona>=1.8 |
| 2 | pandas>=0.24 | |
| 2 | pandas>=0.25 | |
| 3 | 3 | pyproj>=2.2.0 |
| 4 | 4 | shapely>=1.6 |
| 5 | 5 | |
| 14 | 14 | matplotlib>=2.2 |
| 15 | 15 | mapclassify |
| 16 | 16 | |
| 17 | # testing | |
| 17 | # testing | |
| 18 | 18 | pytest>=3.1.0 |
| 19 | 19 | pytest-cov |
| 20 | 20 | codecov |
| 21 | 21 | |
| 22 | # spatial access methods | |
| 22 | # spatial access methods | |
| 23 | 23 | rtree>=0.8 |
| 24 | 24 | |
| 25 | 25 | # styling |
| 29 | 29 | INSTALL_REQUIRES = [] |
| 30 | 30 | else: |
| 31 | 31 | INSTALL_REQUIRES = [ |
| 32 | "pandas >= 0.24.0", | |
| 32 | "pandas >= 0.25.0", | |
| 33 | 33 | "shapely >= 1.6", |
| 34 | 34 | "fiona >= 1.8", |
| 35 | 35 | "pyproj >= 2.2.0", |
| 66 | 66 | "geopandas.tools.tests", |
| 67 | 67 | ], |
| 68 | 68 | package_data={"geopandas": data_files}, |
| 69 | python_requires=">=3.6", | |
| 69 | python_requires=">=3.7", | |
| 70 | 70 | install_requires=INSTALL_REQUIRES, |
| 71 | 71 | cmdclass=versioneer.get_cmdclass(), |
| 72 | 72 | ) |